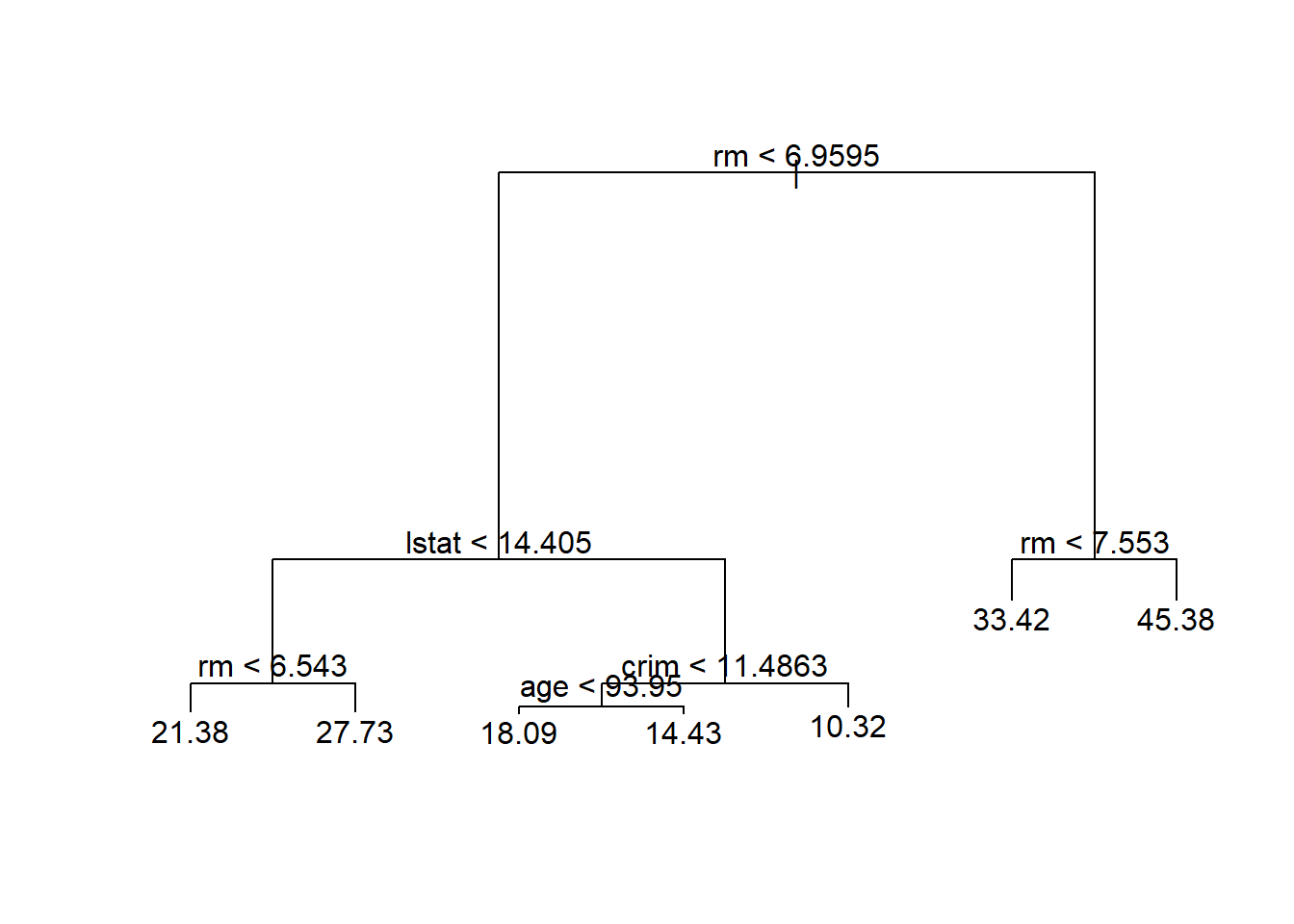16 树方法
调入需要的扩展包:
## Loading required package: Matrix## Loaded glmnet 4.1-8## randomForest 4.7-1.2## Type rfNews() to see new features/changes/bug fixes.## Loaded gbm 2.2.2## This version of gbm is no longer under development. Consider transitioning to gbm3, https://github.com/gbm-developers/gbm316.1 决策树
决策树方法按不同自变量的不同值, 分层地把训练集分组。 每层使用一个变量, 所以这样的分组构成一个二叉树表示。 为了预测一个观测的类归属, 找到它所属的组, 用组的类归属或大多数观测的类归属进行预测。 这样的方法称为决策树(decision tree)。 决策树方法既可以用于判别问题, 也可以用于回归问题。
决策树的好处是容易解释, 在自变量为分类变量时没有额外困难。 但预测准确率可能比其它有监督学习方法差。
改进方法包括装袋法(bagging)、随机森林(random forests)、 提升法(boosting)。 这些改进方法都是把许多棵树合并在一起, 通常能改善准确率但是可解释性变差。
16.1.1 回归树
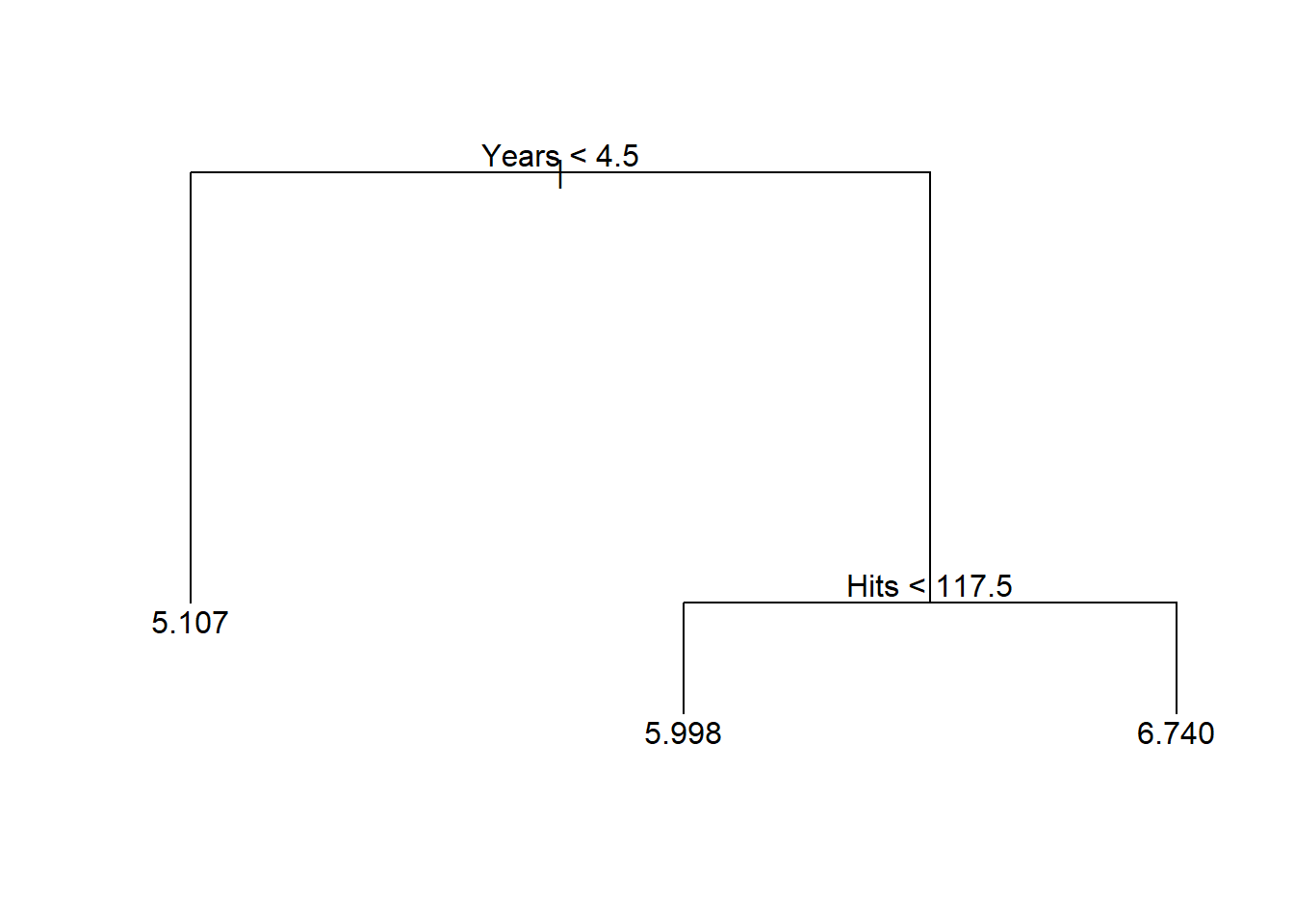
以Hitters数据为例。 以Salary为因变量, 其它自变量为自变量,预测工资值。 为简单起见,仅考虑Years(入行年限)和Hitts(前一年的安打数)两个自变量。 数据需要去掉有缺失值的观测; 以Salary的自然对数值为因变量。 可以得到如下的树(见图16.1):
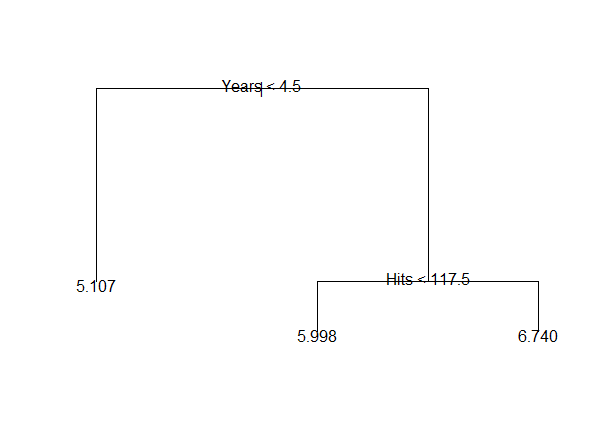
图16.1: Hitters数据的简单树图形
这个树把(Years, Hits)的取值\(\mathbb R^2\)空间分成了三个区域: \[\begin{aligned} R_1 =& \{ (\text{Years}, \text{Hits}):\; \text{Years} < 4.5 \}; \\ R_2 =& \{ (\text{Years}, \text{Hits}):\; \text{Years} \geq 4.5 \text{且} \text{Hits} < 117.5 \}; \\ R_3 =& \{ (\text{Years}, \text{Hits}):\; \text{Years} \geq 4.5 \text{且} \text{Hits} \geq 117.5 \} . \end{aligned}\] 每个区域内用因变量平均值预测因变量值。 见图16.2。
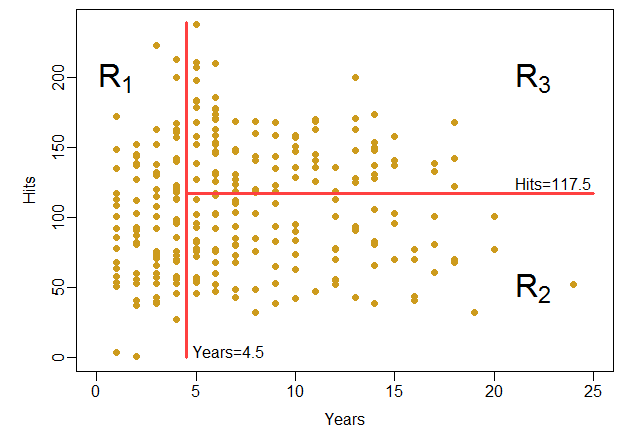
图16.2: Hitters数据的简单树划分的空间
此树对应的文字规则如下:
node), split, n, deviance, yval
* denotes terminal node
1) root 263 207.20 5.927
2) Years < 4.5 90 42.35 5.107 *
3) Years > 4.5 173 72.71 6.354
6) Hits < 117.5 90 28.09 5.998 *
7) Hits > 117.5 83 20.88 6.740 *根据某变量的值分为两个分支的节点称为内部节点; 不再分割的节点称为叶结点。
\(\text{Year}<4.5\)节点和\(\text{Year} \geq 4.5\) 下面的\(\text{Hits}<117.5\)节点是两个内部节点。 \(R_1, R_2, R_3\)是三个叶结点。叶节点处用训练数据的因变量平均值作预测。
从上述结果看出, 为了预测Salary,Years是比Hits更重要的变量。 入行年限多的运动员工资高。 在入行年限超过4.5年的运动员中, Hits低的运动员工资比入行年限低于4.5年的运动员工资提高有限, 而Hits高的运动员工资则提高很多。
树模型可能会过于简单, 但是容易解释和绘图。
树回归步骤用变量值作为分界, 把自变量取值空间\(\mathbb R^p\)分割为不相交的\(J\)个区域 \(R_1, R_2, \dots, R_J\)。 对每个待预测观测,如果自变量值落入\(R_j\)中, 则因变量值用\(R_j\)中训练集的因变量平均值来预测。
16.1.1.1 树回归的分叉方法
树回归分割的区域都是高维矩形或盒子。 求盒子\(R_1, \dots, R_J\)使得其并集为自变量取值空间, 且使得如下的残差平方和最小: \[\begin{align} \text{RSS} = \sum_{j=1}^J \sum_{i \in R_j} (y_i - \hat y_{R_j})^2, \tag{16.1} \end{align}\] 其中\(\hat y_{R_j}\)是\(R_j\)中的训练样本的因变量平均值。
为此,使用由顶向下、贪婪的递归二分叉方法。
由顶向下: 从所有训练样本都在一类中开始,逐步分割, 每次按照一个自变量的值把一个类分成两个类。
贪婪是指每次分割时仅考虑当前使得残差平方和减少最多的分割, 不考虑会对整棵树的影响。
在所有观测都未分割时,要找到一个变量\(X_j\)和变量值\(s_j\), 使得 \[\begin{aligned} R_1(j, s_j) =\{ \boldsymbol x | x_j < s_j \}, \quad R_2(j, s_j) =\{ \boldsymbol x | x_j \geq s_j \} \end{aligned}\] 对应的RSS总和最小, 对每个变量都测试一遍, 找到分割后分割的两个区域内的RSS总和最小的变量和分割点。
对变量\(X_j\),为了求\(s_j\),只要对\(X_j\)的所有不同样本值穷举即可。
在已经分成两类\(R_1\), \(R_2\)以后, 对每一类分别去找到能进行最优分割的变量和截断值\(s\), 然后仅取使得RSS总和更小的那个。 这样得到三个类。 对每个类都找最优分割, 然后仅取使得RSS总和最小的一个, 得到四个类。 如此重复,直到满足某种停止法则, 比如每个类中因变量值都相等, 每个类的观测个数至多5个等等。 分割结束后,对测试样本可以用自变量所属区域的因变量平均值预测因变量值。
16.1.1.2 剪枝
分类树可能会过于复杂, 也会产生过度拟合。 剪去一些枝叶, 使得模型复杂度降低, 可以提高可解释性, 降低方差, 当然偏差会增大一些, 总的来说适当剪枝可以降低测试集上的均方误差。
虽然可以用提前结束分割的方法降低复杂度, 但是这样可能会漏掉底层的好分割。 所以还是先构造一棵大树再剪枝更好。
剪枝的目标是使得测试均方误差最小。 可以用交叉验证或者测试集来估计不同的子树的均方误差。 但是对所有可能的子树都估计均方误差带来过多的计算量。 应仅考虑部分子树。
剪枝的一种方法是代价复杂度剪枝(cost complexity pruning), 也称为最弱联系剪枝(weakest link pruning)。 仅考虑用一个调节参数\(\alpha\)标记的子树的序列。 对每个\(\alpha>0\), 存在子树\(T\), 使得如下函数最小化: \[\begin{aligned} \sum_{m=1}^{|T|} \sum_{i:\; x_i \in R_m} (y_i - \hat y_{R_m})^2 + \alpha |T| \end{aligned}\] 其中\(|T|\)是子树\(T\)的叶子节点个数, \(R_m\)是第\(m\)个叶子节点对应的分割区域, \(\hat y_{R_m}\)是用\(R_m\)中样品计算的因变量预测值。
\(\alpha=0\)时对应于不剪枝的树。 \(\alpha\)增大时规则对较大的树有惩罚, 所以能选择较小的子树, 而且每次子树变化都是从前面较小的\(\alpha\)对应的子树中剪去一个分枝点, 是嵌套进行的。
很容易得到所有的\(\alpha\)对应的子树序列。 可以用交叉验证或测试集选择合适的\(\alpha\)从而选择合适的子树。 得到最优的\(\alpha\)后, 可以再利用全部观测以及获得的\(\alpha\)值产生剪枝的树。
16.1.2 Hitters回归树的简单演示
对Hitters数据,用Years和Hits作因变量预测log(Salaray)。
仅取Hitters数据集的Salary, Years, Hits三个变量, 并仅保留完全观测:
## 'data.frame': 263 obs. of 3 variables:
## $ Salary: num 475 480 500 91.5 750 ...
## $ Years : int 14 3 11 2 11 2 3 2 13 10 ...
## $ Hits : int 81 130 141 87 169 37 73 81 92 159 ...
## - attr(*, "na.action")= 'omit' Named int [1:59] 1 16 19 23 31 33 37 39 40 42 ...
## ..- attr(*, "names")= chr [1:59] "-Andy Allanson" "-Billy Beane" "-Bruce Bochte" "-Bob Boone" ...
## NULL建立完整的树:
剪枝为只有3个叶结点:
显示树:
## node), split, n, deviance, yval
## * denotes terminal node
##
## 1) root 263 207.20 5.927
## 2) Years < 4.5 90 42.35 5.107 *
## 3) Years > 4.5 173 72.71 6.354
## 6) Hits < 117.5 90 28.09 5.998 *
## 7) Hits > 117.5 83 20.88 6.740 *显示概括:
##
## Regression tree:
## snip.tree(tree = tr1, nodes = c(6L, 2L))
## Number of terminal nodes: 3
## Residual mean deviance: 0.3513 = 91.33 / 260
## Distribution of residuals:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -2.24000 -0.39580 -0.03162 0.00000 0.33380 2.55600做树图:
16.1.3 Hitters数据回归树的完整演示
把数据随机地分成一半训练集,一半测试集:
d <- na.omit(Hitters[,c('Salary', 'Years', 'Hits')])
set.seed(1)
train_id <- sample(nrow(d), size=round(0.5*nrow(d)))
train <- rep(FALSE, nrow(d))
train[train_id] <- TRUE
test <- (!train)对训练集,建立未剪枝的树:

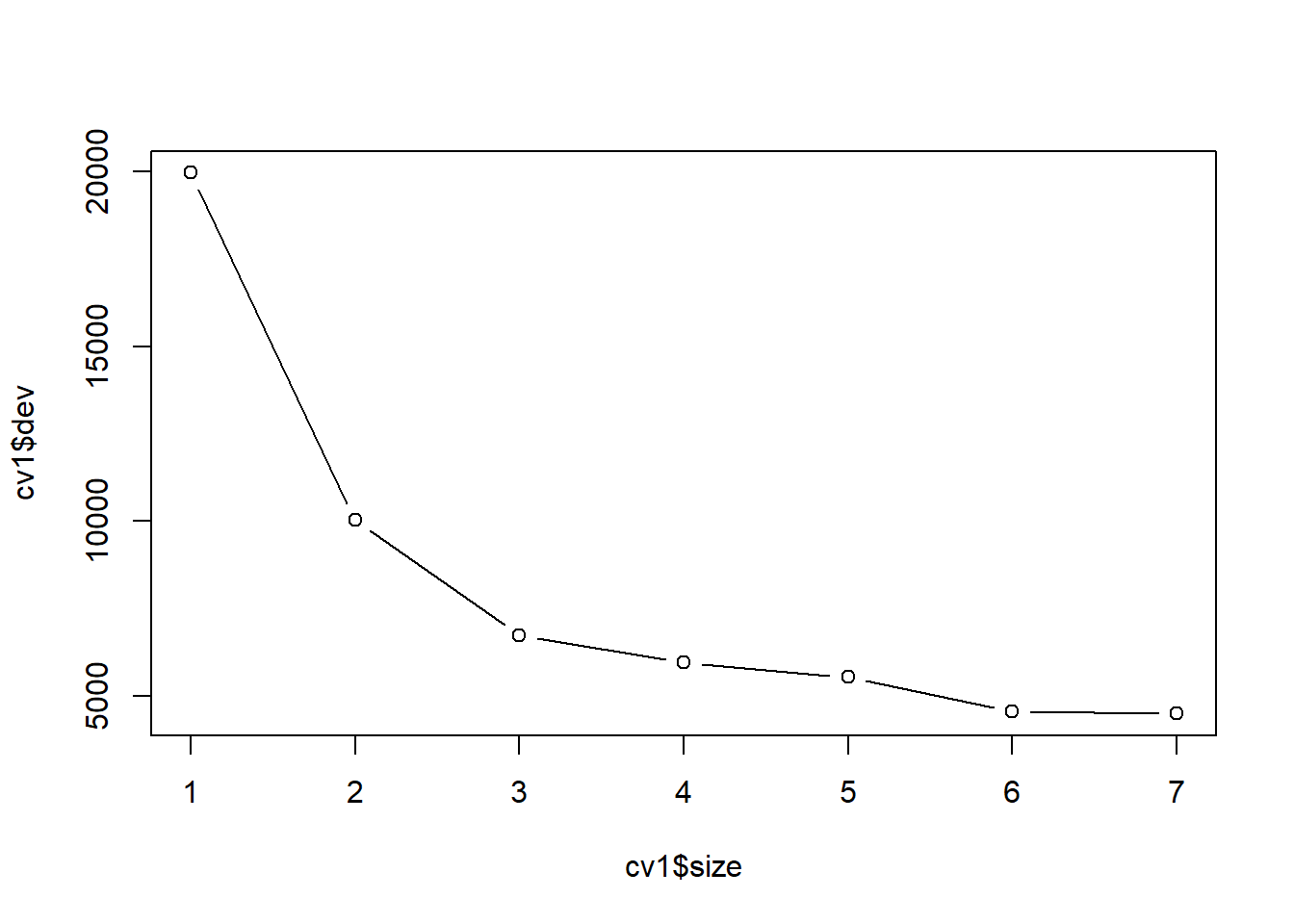
对训练集上的未剪枝树用交叉验证方法寻找最优大小:
## $size
## [1] 7 6 5 4 3 2 1
##
## $dev
## [1] 49.88386 49.78504 50.36061 50.00678 54.07967 66.20924 104.06611
##
## $k
## [1] -Inf 1.558635 1.642895 2.319001 6.275232 11.126931 43.728005
##
## $method
## [1] "deviance"
##
## attr(,"class")
## [1] "prune" "tree.sequence"plot(cv1$size, cv1$dev, type='b')
best.size <- cv1$size[which.min(cv1$dev)[1]]
abline(v=best.size, col='gray')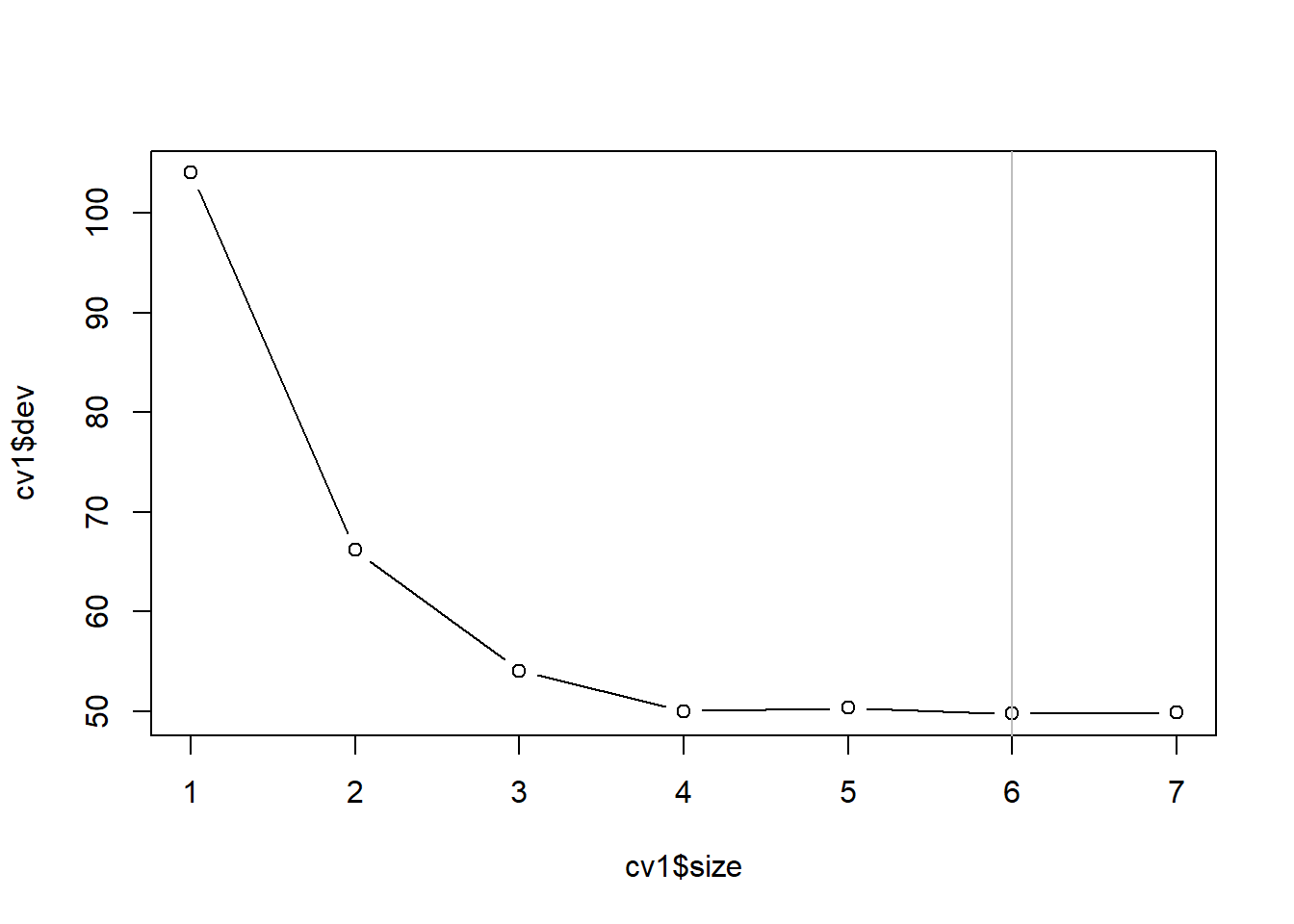
最优大小为6。 获得训练集上构造的树剪枝后的结果:
在测试集上计算预测均方误差:
pred.test <- predict(tr1b, newdata=d[test,])
test.mse <- mean( (d[test, 'Salary'] - exp(pred.test))^2 )
test.mse## [1] 135635.4如果用训练集的因变量平均值估计测试集的因变量值, 均方误差为:
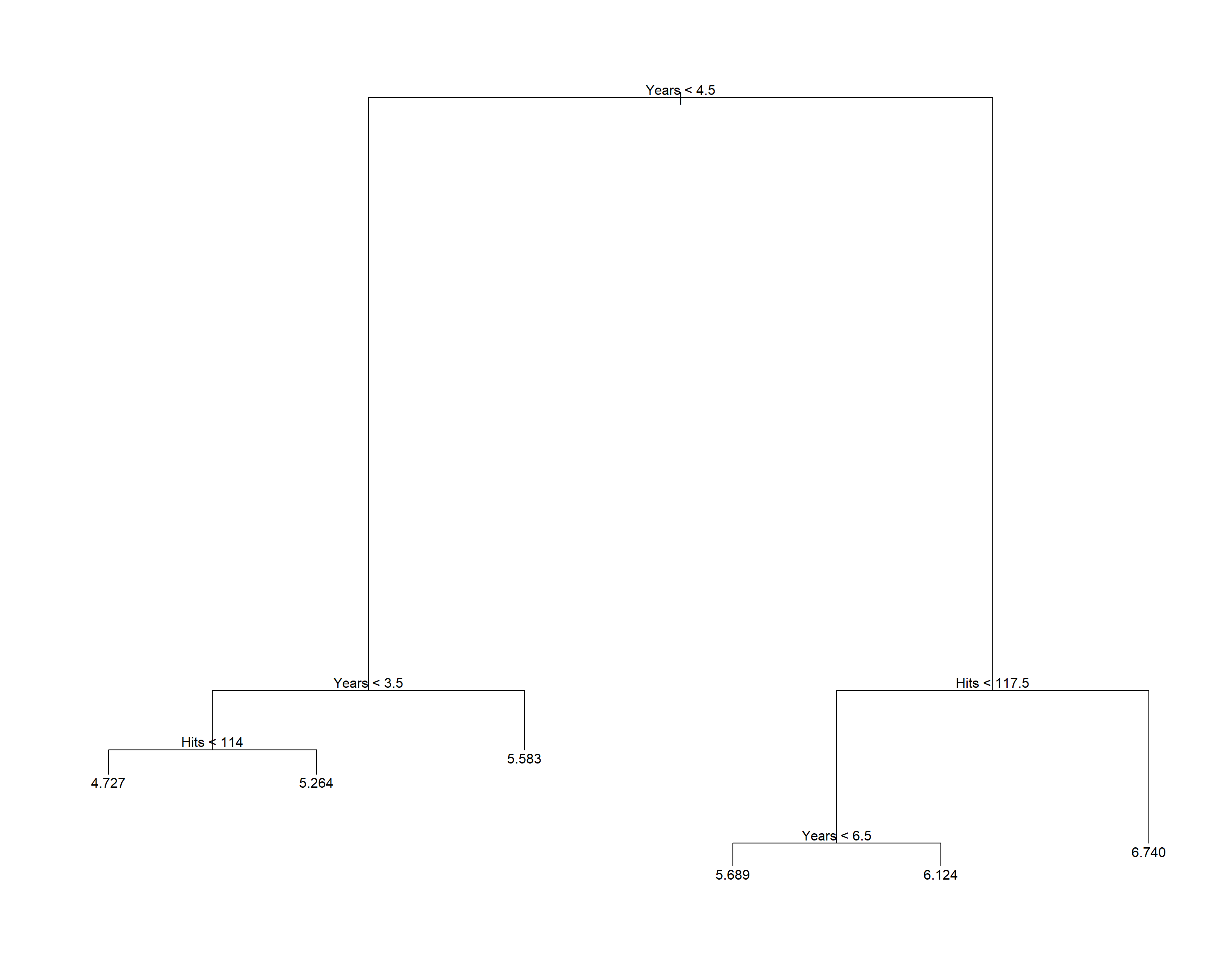
## [1] 224692.1用所有数据来构造未剪枝树:
用训练集上得到的子树大小剪枝:
16.1.4 分类树
这里分类树指因变量是分类变量的树模型。 回归树对每一分割区域用该区域观测的因变量平均值做预测, 而分类树对每一分割区域用该区域因变量的最常见值做预测。 训练出来的回归函数不仅包括区域分割信息、每个区域的最常见类别信息, 还包括每个分割区域的类别分布信息。
在构造分叉时, 不再以拟合均方误差RSS最小为目标, 而是以错判率(classification error rate)最小为目标。 每个区域的错判率是该区域中最常见类之外的类比例。
设\(\hat p_{mk}\)表示第\(m\)个分割区域的训练样本中, 因变量的第\(k\)个类的比例,则 \[\begin{aligned} E = 1 - \max_{k} \hat p_{mk} \end{aligned}\] 是第\(m\)个分割区域的错判率。 但是,错判率不容易区分不同树的判别性能。 一般改用基尼系数和互熵。
基尼系数(Gini index)定义为 \[\begin{aligned} G = \sum_{k=1}^K \hat p_{mk} (1 - \hat p_{mk}), \end{aligned}\] 易见当比例\(\hat p_{mk}\)中一个接近于1,其余都接近于0时\(G\)很小, 这时区域\(m\)基本上都属于因变量的同一类别。 所以\(G\)可以看成是某个分割区域的类混杂程度, \(G\)很小时此区域的类是纯一的, \(G\)很大时,此区域的类别比较混杂,应进一步分割。
另一个衡量某分割区域类别分布情况的指标是如下的互熵(cross entropy): \[\begin{aligned} D = - \sum_{k=1}^K \hat p_{mk} \log \hat p_{mk} . \end{aligned}\] 易见\(D \geq 0\), 当比例\(\hat p_{mk}\)中一个接近于1, 其余都接近于0时\(D\)很小, 这时区域\(m\)基本上都属于因变量的同一类别。 所以\(D\)也可以看成是某个分割区域的类混杂程度, \(D\)很小时此区域的类是纯一的, \(D\)很大时,此区域的类别比较混杂,应进一步分割。 事实上,\(G\)与\(D\)的值很接近。
在分叉时, 用基尼系数或者互熵可以得到更好的分叉结果。
在剪枝时, 仍可以使用基尼系数或互熵, 但是如果关心的是剪枝后的树的预测准确率, 用错判率剪枝更合适。
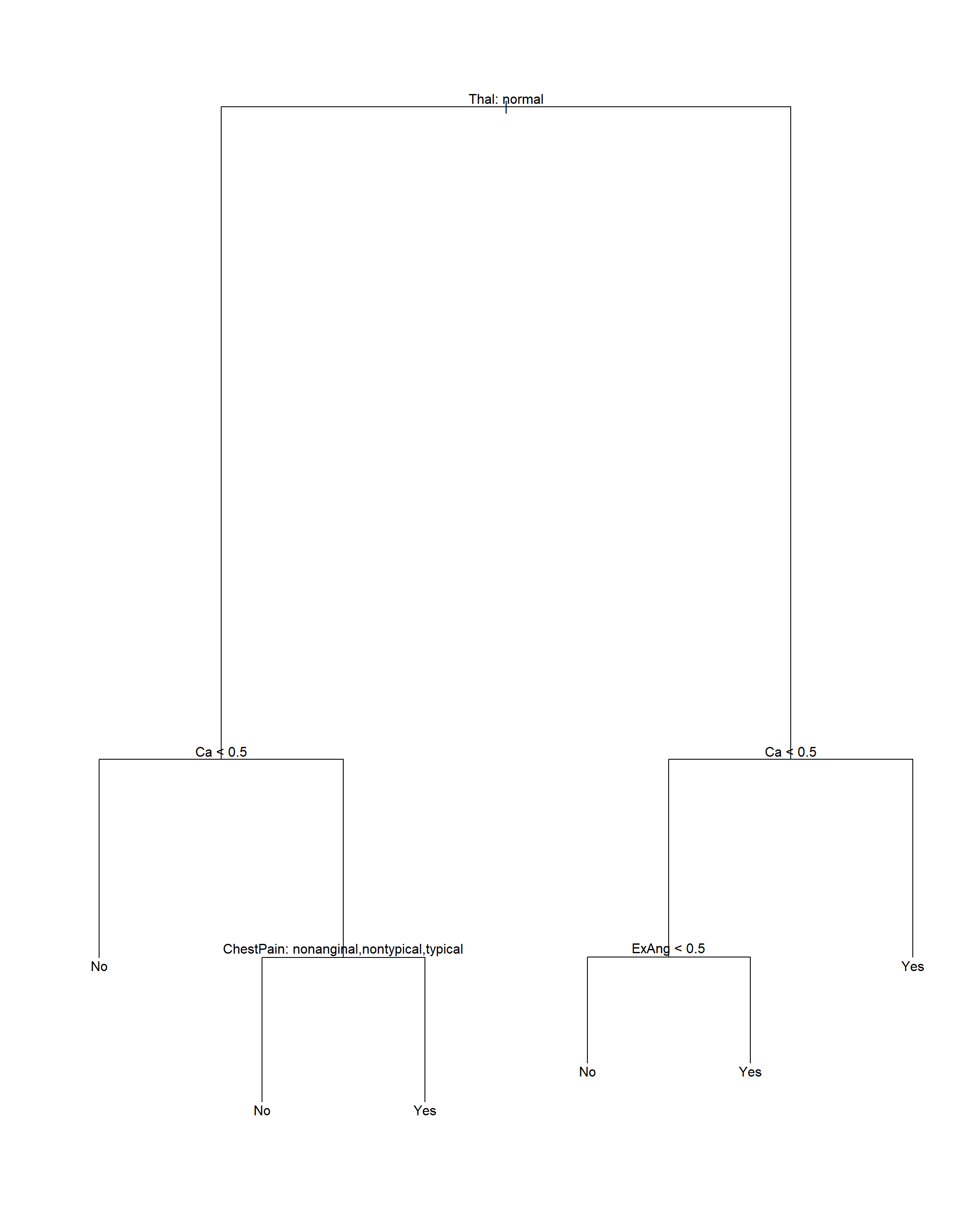
16.1.5 Heart数据判别树演示
数据集Heart中有303个病人的数据,其中变量AHD是二值变量, 取Yes表示用血管造影检查确诊心脏病的,No表示没有心脏病的。 有13个自变量,包括Age, Sex, Chol(胆固醇化验指标)等。
读入Heart数据集,并去掉有缺失值的观测:
Heart <- read.csv("Heart.csv", header=TRUE,
row.names=1,
stringsAsFactors=TRUE)
Heart <- na.omit(Heart)
str(Heart)## 'data.frame': 297 obs. of 14 variables:
## $ Age : int 63 67 67 37 41 56 62 57 63 53 ...
## $ Sex : int 1 1 1 1 0 1 0 0 1 1 ...
## $ ChestPain: Factor w/ 4 levels "asymptomatic",..: 4 1 1 2 3 3 1 1 1 1 ...
## $ RestBP : int 145 160 120 130 130 120 140 120 130 140 ...
## $ Chol : int 233 286 229 250 204 236 268 354 254 203 ...
## $ Fbs : int 1 0 0 0 0 0 0 0 0 1 ...
## $ RestECG : int 2 2 2 0 2 0 2 0 2 2 ...
## $ MaxHR : int 150 108 129 187 172 178 160 163 147 155 ...
## $ ExAng : int 0 1 1 0 0 0 0 1 0 1 ...
## $ Oldpeak : num 2.3 1.5 2.6 3.5 1.4 0.8 3.6 0.6 1.4 3.1 ...
## $ Slope : int 3 2 2 3 1 1 3 1 2 3 ...
## $ Ca : int 0 3 2 0 0 0 2 0 1 0 ...
## $ Thal : Factor w/ 3 levels "fixed","normal",..: 1 2 3 2 2 2 2 2 3 3 ...
## $ AHD : Factor w/ 2 levels "No","Yes": 1 2 2 1 1 1 2 1 2 2 ...
## - attr(*, "na.action")= 'omit' Named int [1:6] 88 167 193 267 288 303
## ..- attr(*, "names")= chr [1:6] "88" "167" "193" "267" ...| Age | Min. :29.00 | 1st Qu.:48.00 | Median :56.00 | Mean :54.54 | 3rd Qu.:61.00 | Max. :77.00 |
| Sex | Min. :0.0000 | 1st Qu.:0.0000 | Median :1.0000 | Mean :0.6768 | 3rd Qu.:1.0000 | Max. :1.0000 |
| ChestPain | asymptomatic:142 | nonanginal : 83 | nontypical : 49 | typical : 23 | NA | NA |
| RestBP | Min. : 94.0 | 1st Qu.:120.0 | Median :130.0 | Mean :131.7 | 3rd Qu.:140.0 | Max. :200.0 |
| Chol | Min. :126.0 | 1st Qu.:211.0 | Median :243.0 | Mean :247.4 | 3rd Qu.:276.0 | Max. :564.0 |
| Fbs | Min. :0.0000 | 1st Qu.:0.0000 | Median :0.0000 | Mean :0.1448 | 3rd Qu.:0.0000 | Max. :1.0000 |
| RestECG | Min. :0.0000 | 1st Qu.:0.0000 | Median :1.0000 | Mean :0.9966 | 3rd Qu.:2.0000 | Max. :2.0000 |
| MaxHR | Min. : 71.0 | 1st Qu.:133.0 | Median :153.0 | Mean :149.6 | 3rd Qu.:166.0 | Max. :202.0 |
| ExAng | Min. :0.0000 | 1st Qu.:0.0000 | Median :0.0000 | Mean :0.3266 | 3rd Qu.:1.0000 | Max. :1.0000 |
| Oldpeak | Min. :0.000 | 1st Qu.:0.000 | Median :0.800 | Mean :1.056 | 3rd Qu.:1.600 | Max. :6.200 |
| Slope | Min. :1.000 | 1st Qu.:1.000 | Median :2.000 | Mean :1.603 | 3rd Qu.:2.000 | Max. :3.000 |
| Ca | Min. :0.0000 | 1st Qu.:0.0000 | Median :0.0000 | Mean :0.6768 | 3rd Qu.:1.0000 | Max. :3.0000 |
| Thal | fixed : 18 | normal :164 | reversable:115 | NA | NA | NA |
| AHD | No :160 | Yes:137 | NA | NA | NA | NA |
16.1.5.1 划分训练集与测试集
简单地把观测分为一半训练集、一半测试集:
set.seed(1)
train_id <- sample(nrow(Heart), size=round(0.5*nrow(Heart)))
train <- rep(FALSE, nrow(Heart))
train[train_id] <- TRUE
test <- (!train)
test.y <- Heart[["AHD"]][test]在训练集上建立未剪枝的判别树:
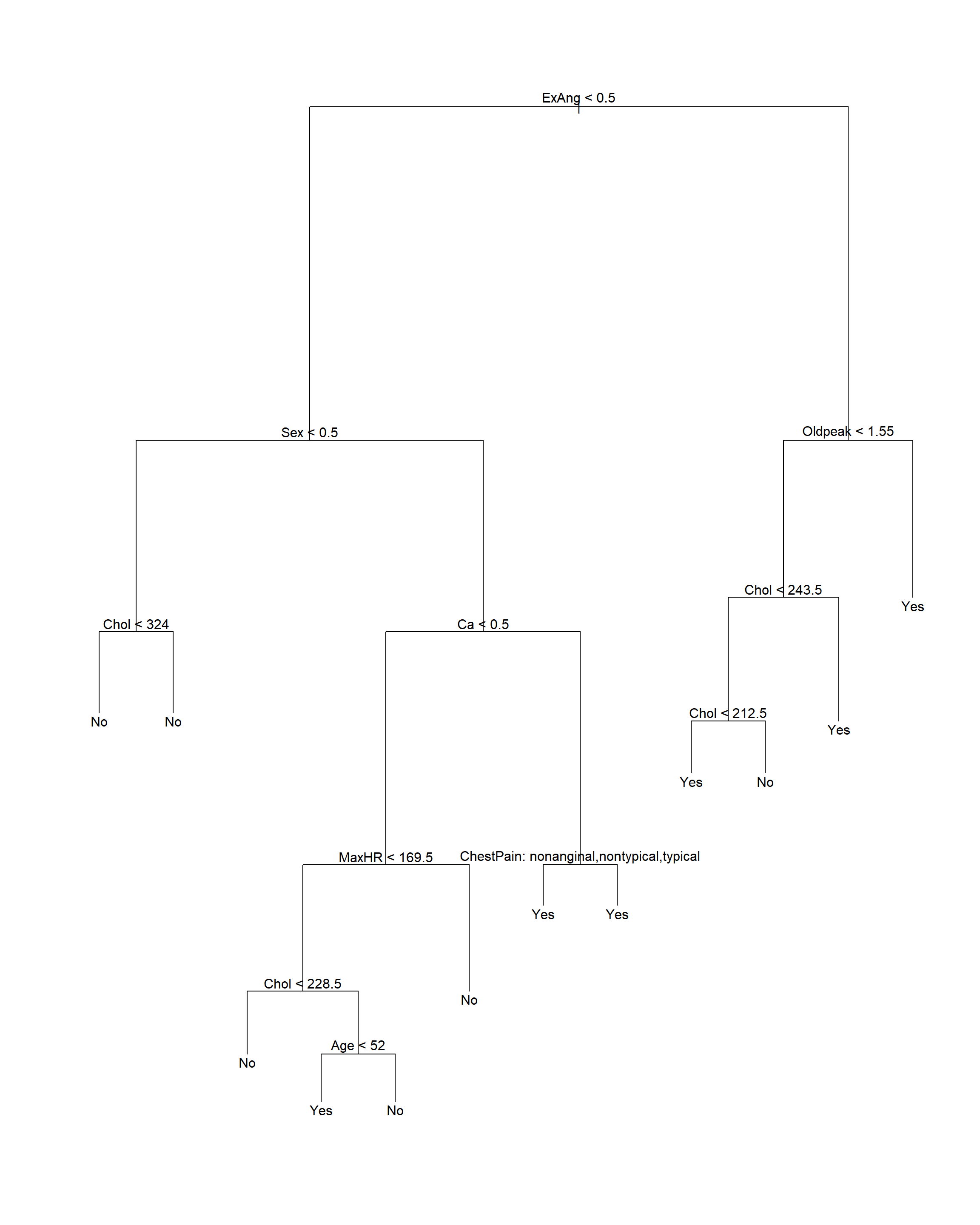
注意: tree()函数要求输入的分类变量为因子类型,
不能直接输入字符型数据作为自变量。
16.1.5.2 适当剪枝
用交叉验证方法确定剪枝保留的叶子个数,剪枝时按照错判率执行:
## $size
## [1] 12 9 6 4 2 1
##
## $dev
## [1] 44 44 40 40 54 66
##
## $k
## [1] -Inf 0.000000 1.666667 3.000000 7.000000 26.000000
##
## $method
## [1] "misclass"
##
## attr(,"class")
## [1] "prune" "tree.sequence"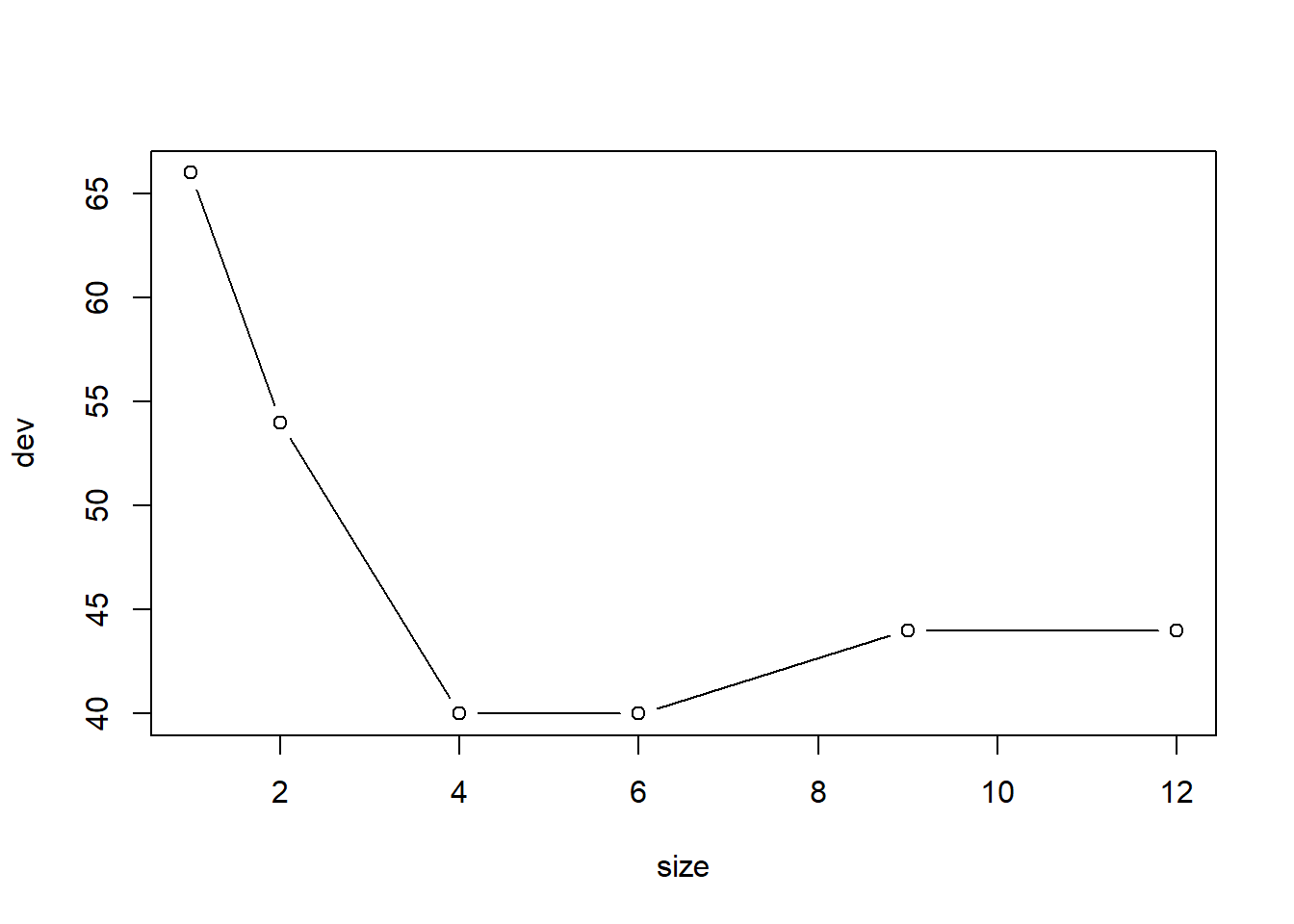
最优的大小是6。
对训练集生成剪枝结果:
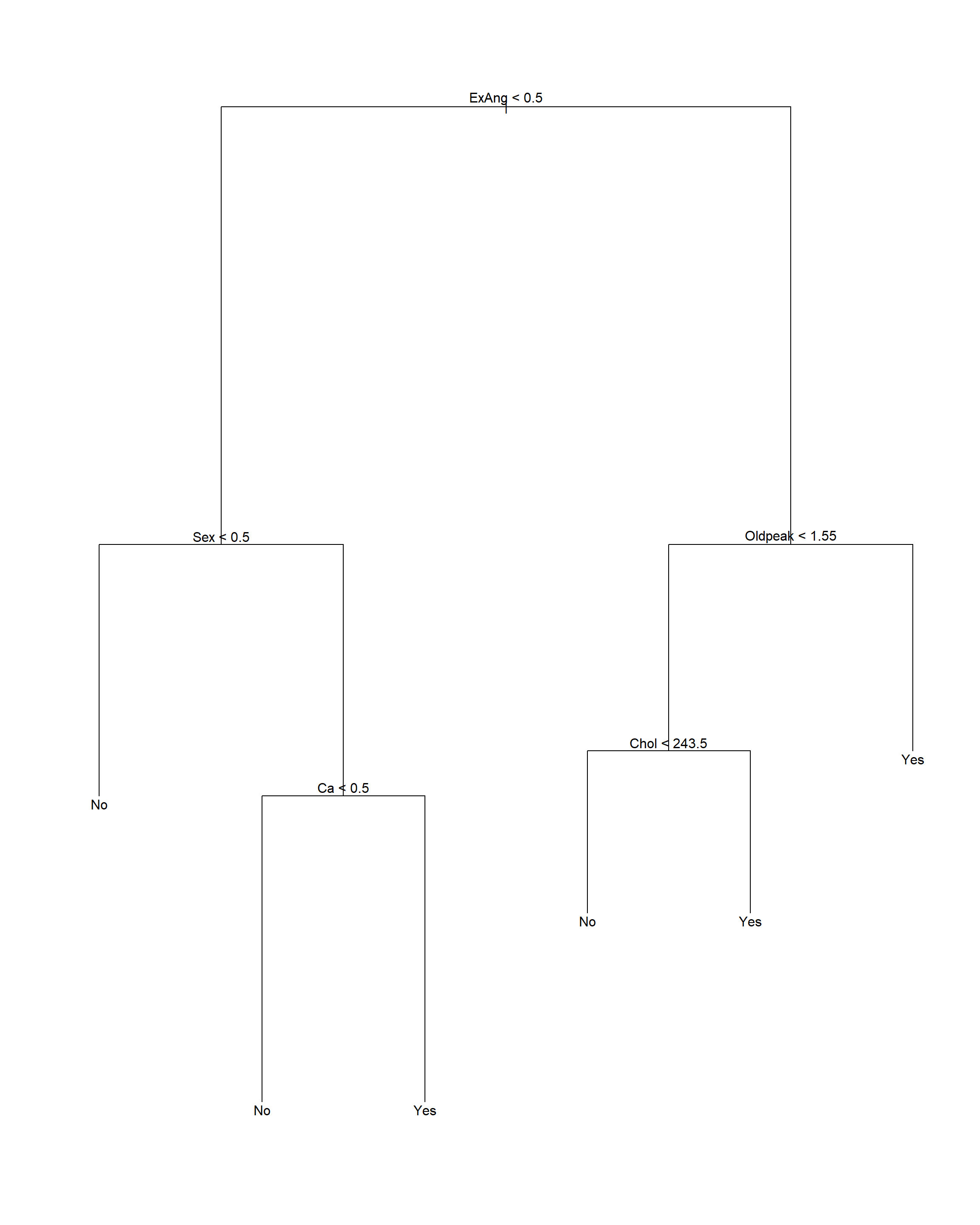
注意剪枝后树的显示中, 内部节点的自变量存在分类变量, 这时按照这个自变量分叉时, 取指定的某几个分类值时对应分支Yes, 取其它的分类值时对应分支No。
16.2 装袋法和随机森林
16.2.1 装袋法
判别树在不同的训练集、测试集划分上可以产生很大变化, 说明其预测值方差较大。 利用bootstrap的思想, 可以随机选取许多个训练集, 把许多个训练集的模型结果平均, 就可以降低预测值的方差。
办法是从一个训练集中用有放回抽样的方法抽取\(B\)个训练集, 设第\(b\)个抽取的训练集得到的回归函数为\(\hat f^{*b}(\cdot)\), 则最后的回归函数是这些回归函数的平均值: \[\begin{aligned} \hat f_{\text{bagging}}(x) = \frac{1}{B} \sum_{b=1}^b \hat f^{*b}(x) \end{aligned}\] 这称为装袋法(bagging)。 装袋法对改善判别与回归树的精度十分有效。
装袋法的步骤如下:
- 从训练集中取\(B\)个有放回随机抽样的bootstrap训练集,\(B\)取为几百到几千之间。
- 对每个bootstrap训练集,估计未剪枝的树。
- 如果因变量是连续变量,对测试样品,用所有的树的预测值的平均值作预测。
- 如果因变量是分类变量,对测试样品,可以用所有树预测类的多数投票决定预测值。
装袋法也可以用来改进其他的回归和判别方法。
装袋后不能再用图形表示,模型可解释性较差。 但是,可以度量自变量在预测中的重要程度。 在回归问题中, 可以计算每个自变量在所有\(B\)个树种平均减少的残差平方和的量, 以此度量其重要度。 在判别问题中, 可以计算每个自变量在所有\(B\)个树种平均减少的基尼系数的量, 以此度量其重要度。
除了可以用测试集、交叉验证方法以外, 还可以使用袋外观测预测误差。 用bootstrap再抽样获得多个训练集时每个bootstrap训练集总会遗漏一些观测, 平均每个bootstrap训练集会遗漏三分之一的观测。 对每个观测,大约有\(B/3\)棵树没有用到此观测, 可以用这些树的预测值平均来预测此观测,得到一个误差估计, 这样得到的均方误差估计或错判率称为袋外观测估计(OOB估计)。 好处是不用很多额外的工作。
16.2.1.1 Hitters数据装袋法演示
对训练集用装袋法:
set.seed(1)
d <- na.omit(Hitters)
train_id <- sample(nrow(d), size=round(0.5*nrow(d)))
train <- rep(FALSE, nrow(d))
train[train_id] <- TRUE
test <- (!train)
bag1 <- randomForest(log(Salary) ~ ., data=d,
subset=train, mtry=ncol(d)-1, importance=TRUE)
bag1##
## Call:
## randomForest(formula = log(Salary) ~ ., data = d, mtry = ncol(d) - 1, importance = TRUE, subset = train)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 19
##
## Mean of squared residuals: 0.2488363
## % Var explained: 68.1注意randomForest()函数实际是随机森林法,
但是当mtry的值取为所有自变量个数时就是装袋法。
对测试集进行预报:
pred2 <- predict(bag1, newdata=d[test,])
test.mse2 <- mean( (d[test, 'Salary'] - exp(pred2))^2 )
test.mse2## [1] 90171.44结果与剪枝过的单课树相近。
在全集上使用装袋法:
##
## Call:
## randomForest(formula = log(Salary) ~ ., data = d, mtry = ncol(d) - 1, importance = TRUE)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 19
##
## Mean of squared residuals: 0.1873377
## % Var explained: 76.22变量的重要度数值和图形: 各变量的重要度数值及其图形:
## %IncMSE IncNodePurity
## AtBat 12.24054066 8.5996425
## Hits 9.47716109 8.0146504
## HmRun 4.19762724 2.1570060
## Runs 7.68565733 3.9542718
## RBI 7.51172719 5.7689615
## Walks 8.34560927 6.8880129
## Years 9.25986753 2.2505365
## CAtBat 26.74337522 79.8379721
## CHits 13.34178783 27.0067167
## CHmRun 6.93413792 3.9886086
## CRuns 14.33934038 32.9637351
## CRBI 15.42782700 10.2720711
## CWalks 7.02126823 4.6201072
## League -1.00937275 0.1668663
## Division 0.03884951 0.2590991
## PutOuts 2.96962100 3.7135650
## Assists 1.06380884 1.7233765
## Errors 1.62377900 1.6439786
## NewLeague 1.42880310 0.3362676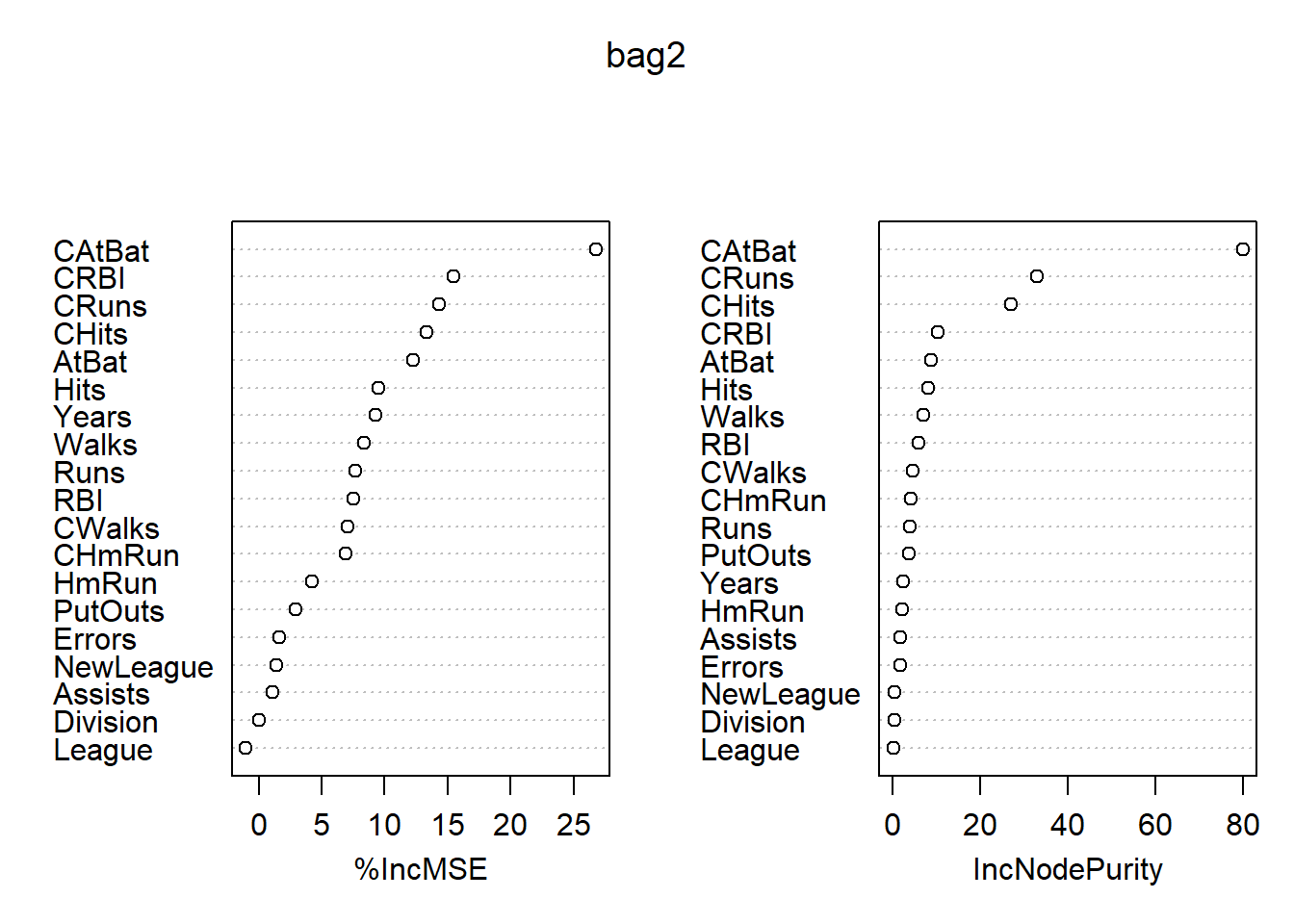
最重要的自变量是CAtBats, 其次有CRuns, CHits, CRBI等。
16.2.1.2 Heart数据用装袋法演示
对训练集用装袋法:
set.seed(1)
train_id <- sample(nrow(Heart), size=round(0.5*nrow(Heart)))
train <- rep(FALSE, nrow(Heart))
train[train_id] <- TRUE
test <- (!train)
test.y <- Heart[["AHD"]][test]
bag1 <- randomForest(AHD ~ ., data=Heart,
subset=train, mtry=13, importance=TRUE)
bag1##
## Call:
## randomForest(formula = AHD ~ ., data = Heart, mtry = 13, importance = TRUE, subset = train)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 13
##
## OOB estimate of error rate: 22.97%
## Confusion matrix:
## No Yes class.error
## No 70 13 0.1566265
## Yes 21 44 0.3230769注意randomForest()函数实际是随机森林法,
但是当mtry的值取为所有自变量个数时就是装袋法。
袋外观测得到的错判率比较差。
对测试集进行预报:
## test.y
## pred2 No Yes
## No 66 16
## Yes 11 56## [1] 0.1812081测试集的错判率约为18%。
对全集用装袋法:
##
## Call:
## randomForest(formula = AHD ~ ., data = Heart, mtry = 13, importance = TRUE)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 13
##
## OOB estimate of error rate: 19.53%
## Confusion matrix:
## No Yes class.error
## No 134 26 0.1625000
## Yes 32 105 0.2335766各变量的重要度数值及其图形:
## No Yes MeanDecreaseAccuracy MeanDecreaseGini
## Age 7.5914270 5.9300895 9.5901487 12.423534
## Sex 11.1035002 4.4224309 12.5552921 3.606976
## ChestPain 11.8110968 17.8134383 20.3909869 23.737158
## RestBP 4.2851096 2.6043117 4.9076443 9.865693
## Chol -1.9750832 -3.4792229 -3.6808119 10.838036
## Fbs 0.7931423 -2.7452667 -1.2269553 0.874251
## RestECG -1.7922866 0.8798907 -0.7548197 1.895233
## MaxHR 8.5471129 1.4362163 7.8503704 13.396300
## ExAng 1.8258307 5.7732467 5.2672188 3.941524
## Oldpeak 15.2223889 13.0068694 19.4903808 14.709831
## Slope 2.9181877 5.2329194 5.8296096 4.200541
## Ca 24.3509148 18.4287501 29.6110614 20.004071
## Thal 21.0348018 18.0554814 26.3633854 27.572038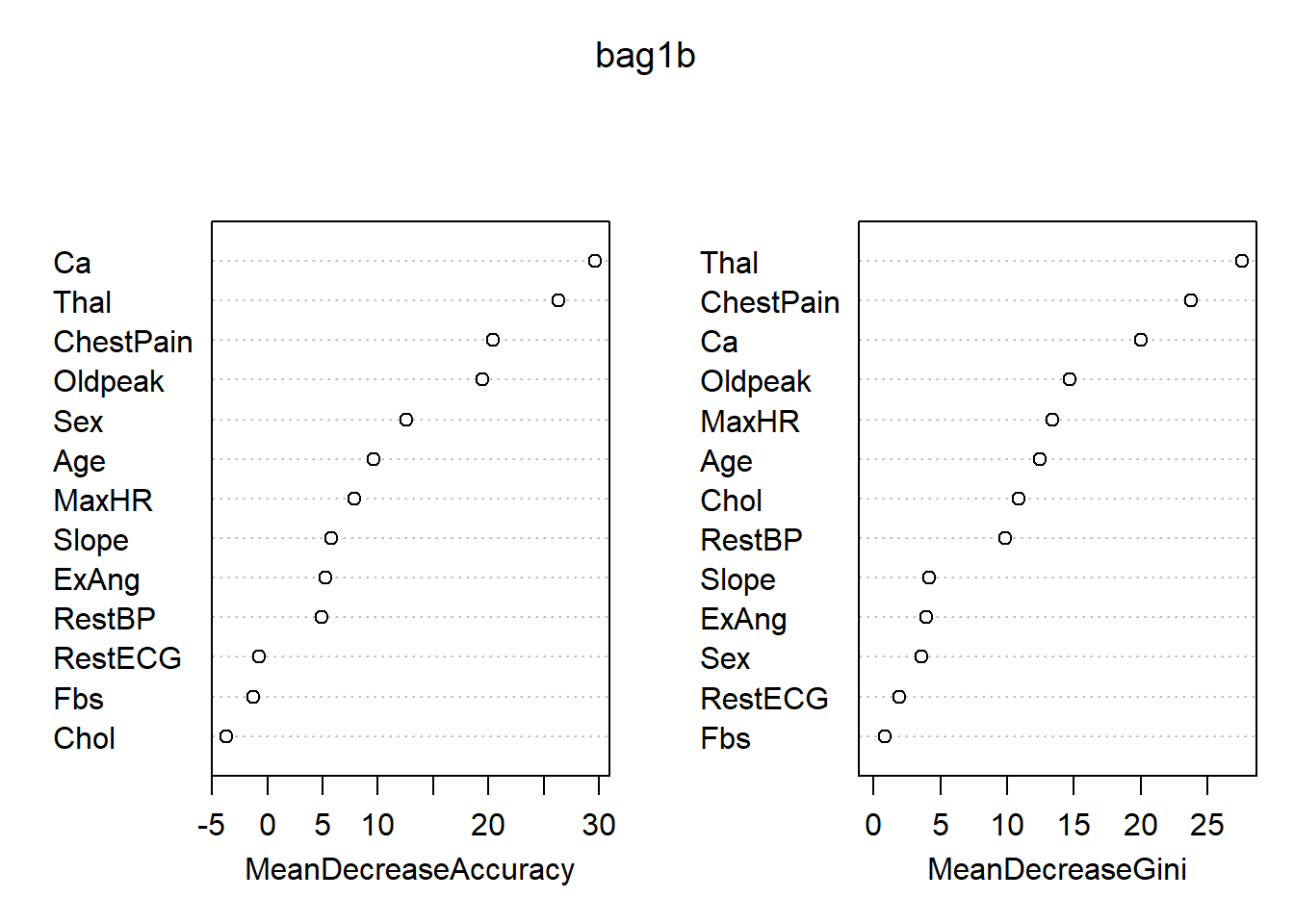
最重要的变量是Thal, ChestPain, Ca。
16.2.2 随机森林
随机森林的思想与装袋法类似, 但是试图使得参加平均的各个树之间变得比较独立。 仍采用有放回抽样得到的多个bootstrap训练集, 但是对每个bootstrap训练集构造判别树时, 每次分叉时不考虑所有自变量, 而是仅考虑随机选取的一个自变量子集。
对判别树,每次分叉时选取的自变量个数通常取\(m \approx \sqrt{p}\)个。 比如,对Heart数据的13个自变量, 每次分叉时仅随机选取4个纳入考察范围。
随机森林的想法是基于正相关的样本在平均时并不能很好地降低方差, 独立样本能比较好地降低方差。 如果存在一个最重要的变量, 如果不加限制这个最重要的变量总会是第一个分叉, 使得\(B\)棵树相似程度很高。 随机森林解决这个问题的办法是限制分叉时可选的变量子集。
随机森林也可以用来改进其他的回归和判别方法。
装袋法和随机森林都可以用R扩展包randomForest的
randomForest()函数实现。
当此函数的mtry参数取为自变量个数时,执行的就是装袋法;
mtry取缺省值时,执行随机森林算法。
执行随机森林算法时,
randomForest()函数在回归问题时分叉时考虑的自变量个数取\(m \approx p/3\),
在判别问题时取\(m \approx \sqrt{p}\)。
16.2.2.1 Hitters数据随机森林演示
对训练集用随机森林法:
set.seed(1)
d <- na.omit(Hitters)
train_id <- sample(nrow(d), size=round(0.5*nrow(d)))
train <- rep(FALSE, nrow(d))
train[train_id] <- TRUE
test <- (!train)
rf1 <- randomForest(log(Salary) ~ ., data=d,
subset=train, importance=TRUE)
rf1##
## Call:
## randomForest(formula = log(Salary) ~ ., data = d, importance = TRUE, subset = train)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 6
##
## Mean of squared residuals: 0.2437535
## % Var explained: 68.75当mtry的值取为缺省值时执行随机森林算法。
对测试集进行预报:
pred3 <- predict(rf1, newdata=d[test,])
test.mse3 <- mean( (d[test, 'Salary'] - exp(pred3))^2 )
test.mse3## [1] 96229.2结果与剪枝过的单课树、装袋法相近。
在全集上使用随机森林:
##
## Call:
## randomForest(formula = log(Salary) ~ ., data = d, importance = TRUE)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 6
##
## Mean of squared residuals: 0.1782609
## % Var explained: 77.37各变量的重要度数值及其图形:
## %IncMSE IncNodePurity
## AtBat 11.81948475 8.2308776
## Hits 8.09787840 7.5075741
## HmRun 5.91570003 2.4320892
## Runs 8.48169025 4.8333805
## RBI 6.29673815 6.3026314
## Walks 9.36982692 6.2217510
## Years 10.61587730 6.5702146
## CAtBat 17.58354435 40.7827117
## CHits 15.64594781 34.4452277
## CHmRun 7.04523612 6.4528435
## CRuns 15.85901741 35.5844665
## CRBI 13.56732483 18.8060878
## CWalks 10.25381701 17.6924487
## League -0.44093551 0.3050351
## Division 0.32697628 0.2715000
## PutOuts 3.74477178 3.0747050
## Assists -0.43394356 1.6667284
## Errors 1.58044791 1.7246783
## NewLeague -0.01057482 0.3535943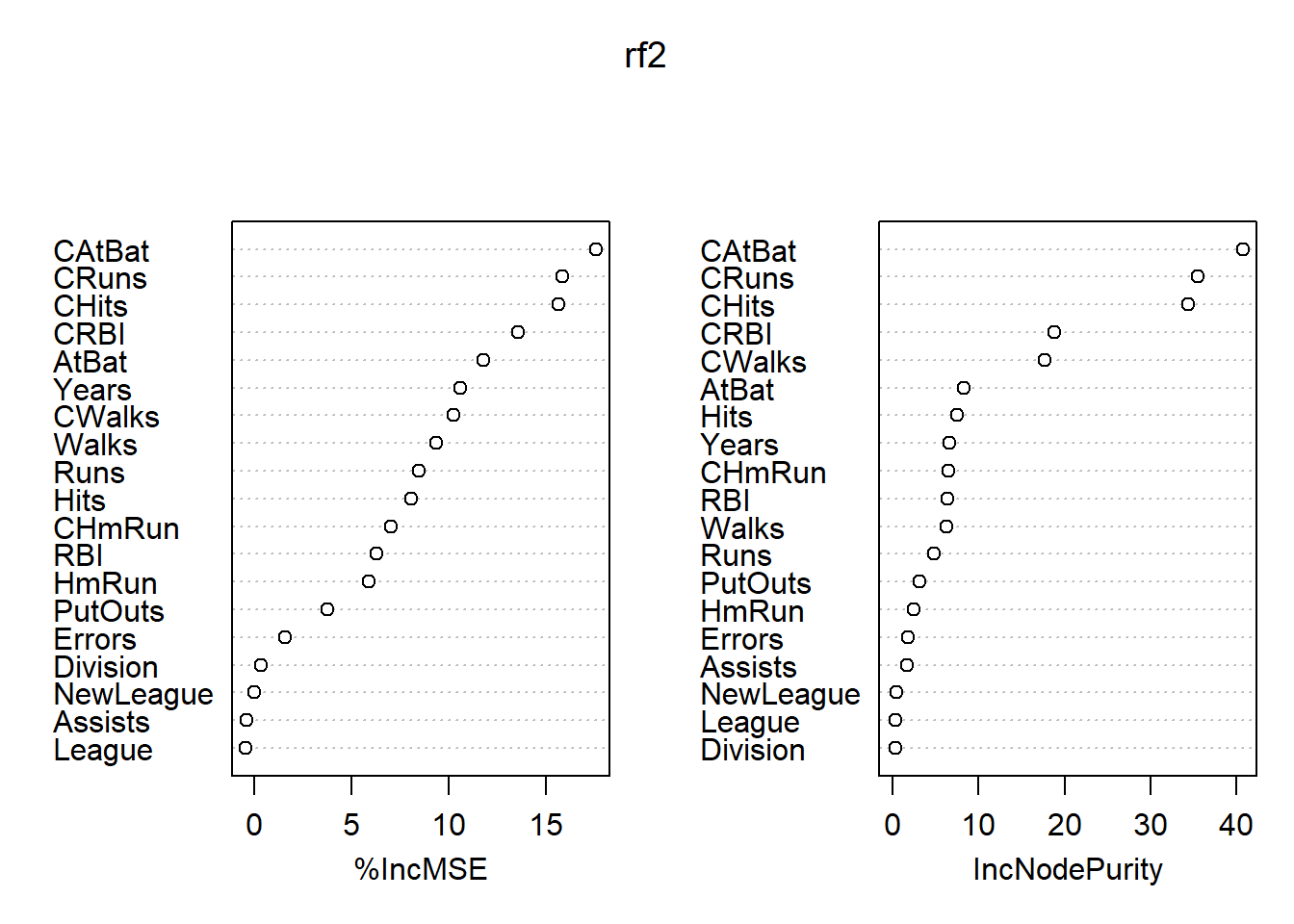
最重要的自变量是CAtBats, CRuns, CHits, CWalks, CRBI等。
16.2.2.2 Heart数据随机森林演示
对训练集用随机森林法:
set.seed(1)
train_id <- sample(nrow(Heart), size=round(0.5*nrow(Heart)))
train <- rep(FALSE, nrow(Heart))
train[train_id] <- TRUE
test <- (!train)
test.y <- Heart[["AHD"]][test]
rf1 <- randomForest(AHD ~ ., data=Heart, subset=train, importance=TRUE)
rf1##
## Call:
## randomForest(formula = AHD ~ ., data = Heart, importance = TRUE, subset = train)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 3
##
## OOB estimate of error rate: 22.97%
## Confusion matrix:
## No Yes class.error
## No 71 12 0.1445783
## Yes 22 43 0.3384615这里mtry取缺省值,对应于随机森林法。
对测试集进行预报:
## test.y
## pred3 No Yes
## No 70 17
## Yes 7 55## [1] 0.1610738测试集的错判率约为16%。
对全集用随机森林:
##
## Call:
## randomForest(formula = AHD ~ ., data = Heart, importance = TRUE)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 3
##
## OOB estimate of error rate: 17.17%
## Confusion matrix:
## No Yes class.error
## No 137 23 0.1437500
## Yes 28 109 0.2043796各变量的重要度数值及其图形:
## No Yes MeanDecreaseAccuracy MeanDecreaseGini
## Age 5.0451528 4.1243916 6.2544913 12.589891
## Sex 11.8830005 5.7666976 13.0527236 4.348853
## ChestPain 12.9853182 15.9344889 20.0063334 18.140518
## RestBP 2.3860139 0.8161766 2.2560609 10.402834
## Chol 0.4441293 -1.7627423 -1.0295881 11.550493
## Fbs 1.3711348 -2.5626917 -0.8151845 1.329501
## RestECG 0.1510264 0.5408153 0.4091251 2.734730
## MaxHR 9.6540260 4.8456013 10.0807989 17.028765
## ExAng 3.8100638 7.2834823 8.0332494 7.365512
## Oldpeak 9.9336236 12.8188739 16.1137642 15.876015
## Slope 1.9767505 8.3713104 7.6713543 6.421739
## Ca 21.6441820 18.4225083 26.1245597 18.106245
## Thal 19.6662985 16.5929688 24.6866916 19.282387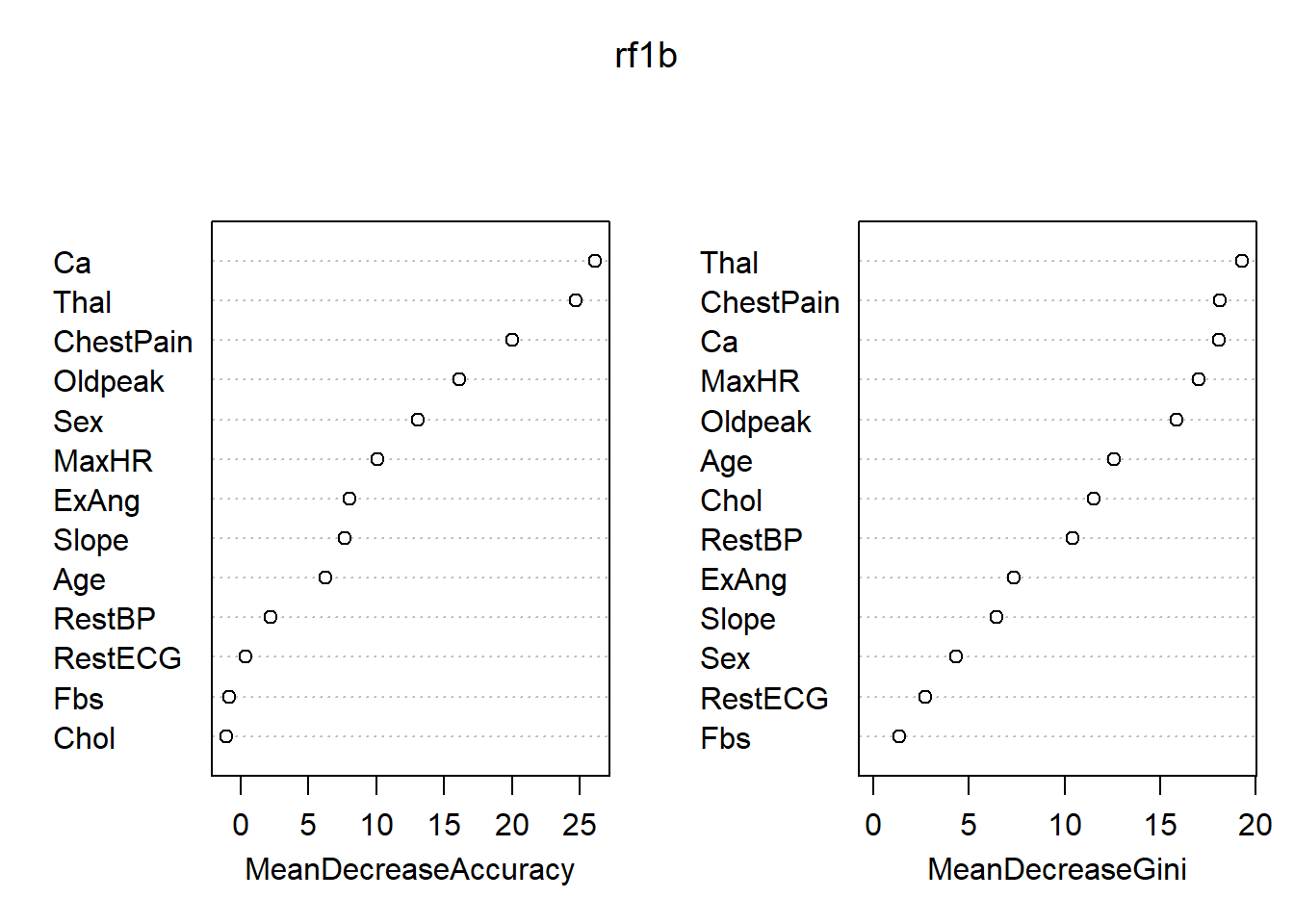
最重要的变量是ChestPain, Thal, Ca。
16.3 汽车销量数据的演示
Carseats是ISLR包的一个数据集,基本情况如下:
## 'data.frame': 400 obs. of 11 variables:
## $ Sales : num 9.5 11.22 10.06 7.4 4.15 ...
## $ CompPrice : num 138 111 113 117 141 124 115 136 132 132 ...
## $ Income : num 73 48 35 100 64 113 105 81 110 113 ...
## $ Advertising: num 11 16 10 4 3 13 0 15 0 0 ...
## $ Population : num 276 260 269 466 340 501 45 425 108 131 ...
## $ Price : num 120 83 80 97 128 72 108 120 124 124 ...
## $ ShelveLoc : Factor w/ 3 levels "Bad","Good","Medium": 1 2 3 3 1 1 3 2 3 3 ...
## $ Age : num 42 65 59 55 38 78 71 67 76 76 ...
## $ Education : num 17 10 12 14 13 16 15 10 10 17 ...
## $ Urban : Factor w/ 2 levels "No","Yes": 2 2 2 2 2 1 2 2 1 1 ...
## $ US : Factor w/ 2 levels "No","Yes": 2 2 2 2 1 2 1 2 1 2 ...## Sales CompPrice Income Advertising Population Price ShelveLoc Age Education Urban US
## Min. : 0.000 Min. : 77 Min. : 21.00 Min. : 0.000 Min. : 10.0 Min. : 24.0 Bad : 96 Min. :25.00 Min. :10.0 No :118 No :142
## 1st Qu.: 5.390 1st Qu.:115 1st Qu.: 42.75 1st Qu.: 0.000 1st Qu.:139.0 1st Qu.:100.0 Good : 85 1st Qu.:39.75 1st Qu.:12.0 Yes:282 Yes:258
## Median : 7.490 Median :125 Median : 69.00 Median : 5.000 Median :272.0 Median :117.0 Medium:219 Median :54.50 Median :14.0
## Mean : 7.496 Mean :125 Mean : 68.66 Mean : 6.635 Mean :264.8 Mean :115.8 Mean :53.32 Mean :13.9
## 3rd Qu.: 9.320 3rd Qu.:135 3rd Qu.: 91.00 3rd Qu.:12.000 3rd Qu.:398.5 3rd Qu.:131.0 3rd Qu.:66.00 3rd Qu.:16.0
## Max. :16.270 Max. :175 Max. :120.00 Max. :29.000 Max. :509.0 Max. :191.0 Max. :80.00 Max. :18.0把Salses变量按照大于8与否分成两组, 结果存入变量High,以High为因变量作判别分析。
## [1] 400 1216.3.1 判别树
16.3.1.1 全体数据的判别树
对全体数据建立未剪枝的判别树:
##
## Classification tree:
## tree(formula = High ~ . - Sales, data = d)
## Variables actually used in tree construction:
## [1] "ShelveLoc" "Price" "Income" "CompPrice" "Population" "Advertising" "Age" "US"
## Number of terminal nodes: 27
## Residual mean deviance: 0.4575 = 170.7 / 373
## Misclassification error rate: 0.09 = 36 / 400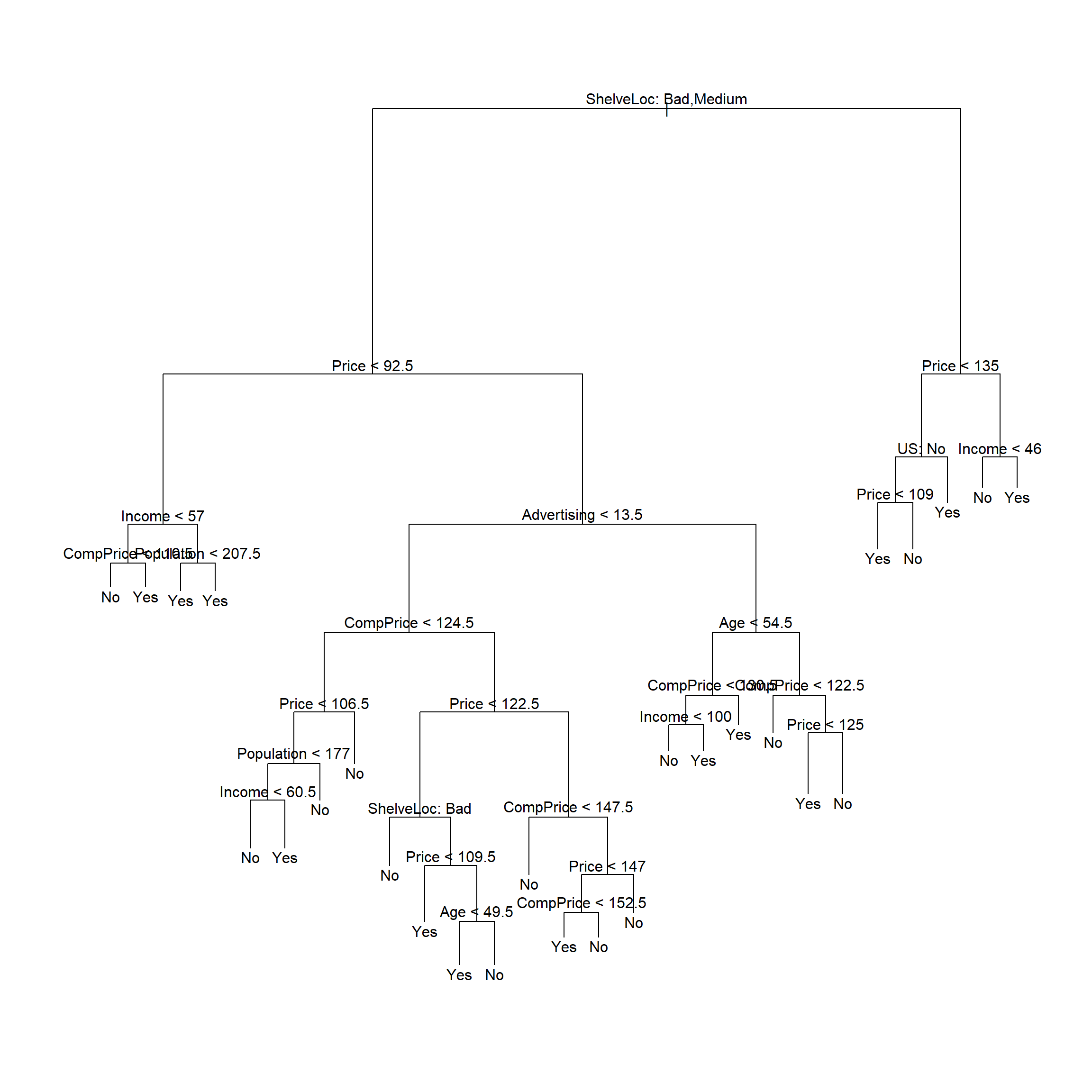
16.3.1.2 划分训练集和测试集
把输入数据集随机地分一半当作训练集,另一半当作测试集:
d <- na.omit(Carseats)
d$High <- factor(ifelse(d$Sales > 8, 'Yes', 'No'))
set.seed(2)
train_id <- sample(nrow(d), size=round(0.5*nrow(d)))
train <- rep(FALSE, nrow(d))
train[train_id] <- TRUE
test <- (!train)
test.high <- d[["High"]][test]用训练数据建立未剪枝的判别树:
##
## Classification tree:
## tree(formula = High ~ . - Sales, data = d, subset = train)
## Variables actually used in tree construction:
## [1] "Price" "Population" "ShelveLoc" "Age" "Education" "CompPrice" "Advertising" "Income" "US"
## Number of terminal nodes: 21
## Residual mean deviance: 0.5543 = 99.22 / 179
## Misclassification error rate: 0.115 = 23 / 200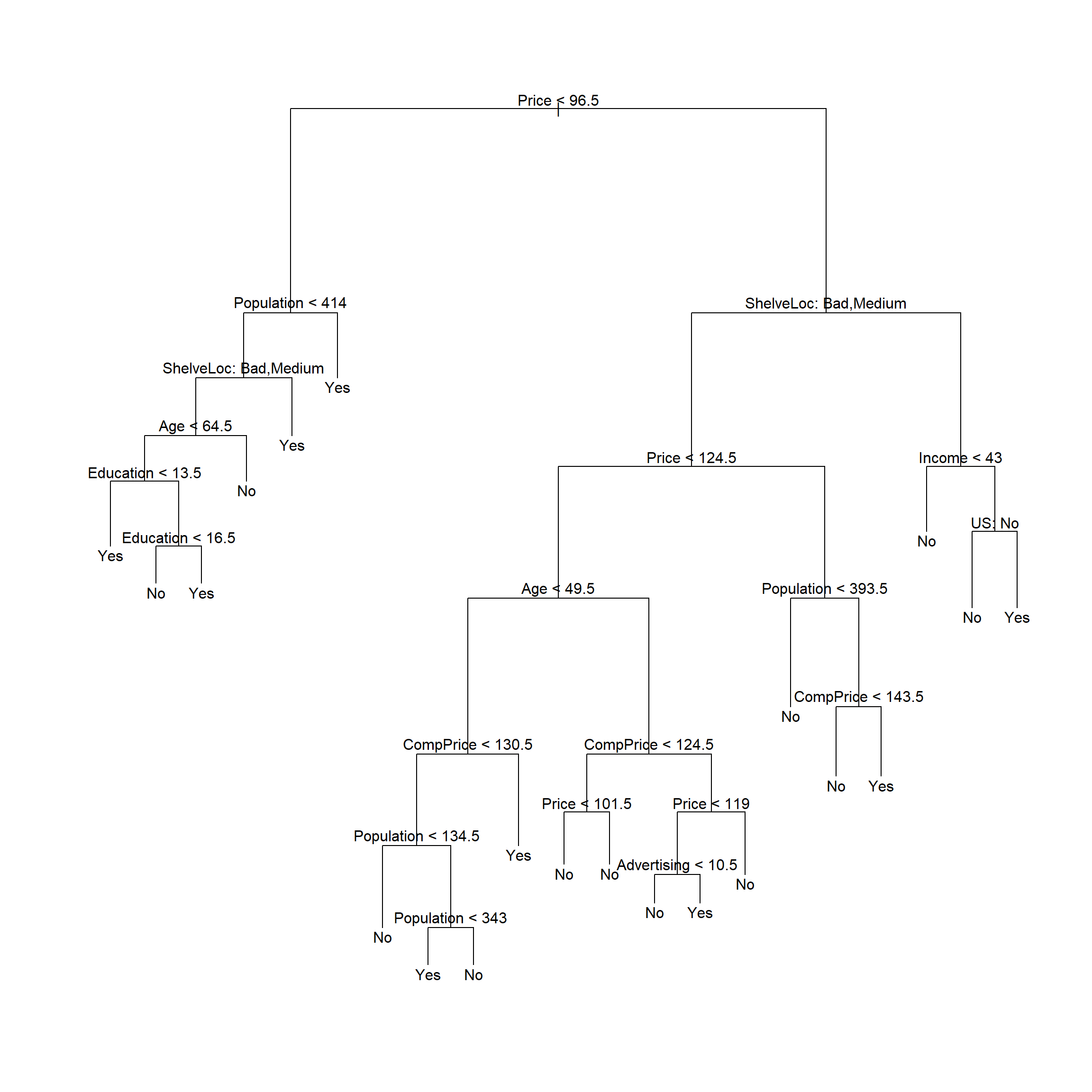
用未剪枝的树对测试集进行预测,并计算误判率:
## test.high
## pred2 No Yes
## No 104 33
## Yes 13 50## [1] 0.2316.3.1.3 用交叉验证确定训练集的剪枝
## $size
## [1] 21 19 14 9 8 5 3 2 1
##
## $dev
## [1] 74 74 78 77 77 79 75 80 82
##
## $k
## [1] -Inf 0.0 1.0 1.4 2.0 3.0 4.0 9.0 18.0
##
## $method
## [1] "misclass"
##
## attr(,"class")
## [1] "prune" "tree.sequence"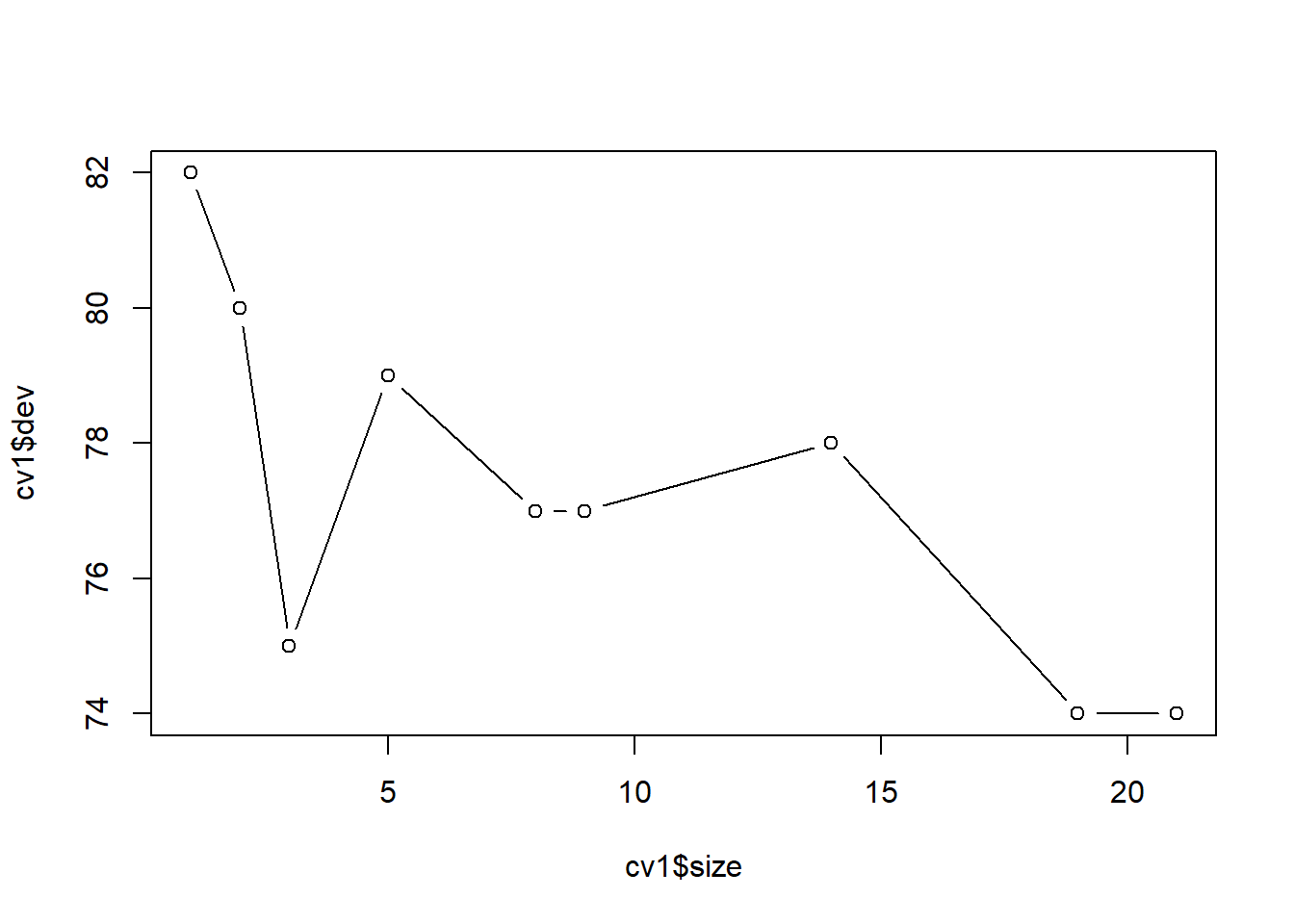
用交叉验证方法自动选择的最佳树大小为21。
剪枝:
##
## Classification tree:
## tree(formula = High ~ . - Sales, data = d, subset = train)
## Variables actually used in tree construction:
## [1] "Price" "Population" "ShelveLoc" "Age" "Education" "CompPrice" "Advertising" "Income" "US"
## Number of terminal nodes: 21
## Residual mean deviance: 0.5543 = 99.22 / 179
## Misclassification error rate: 0.115 = 23 / 200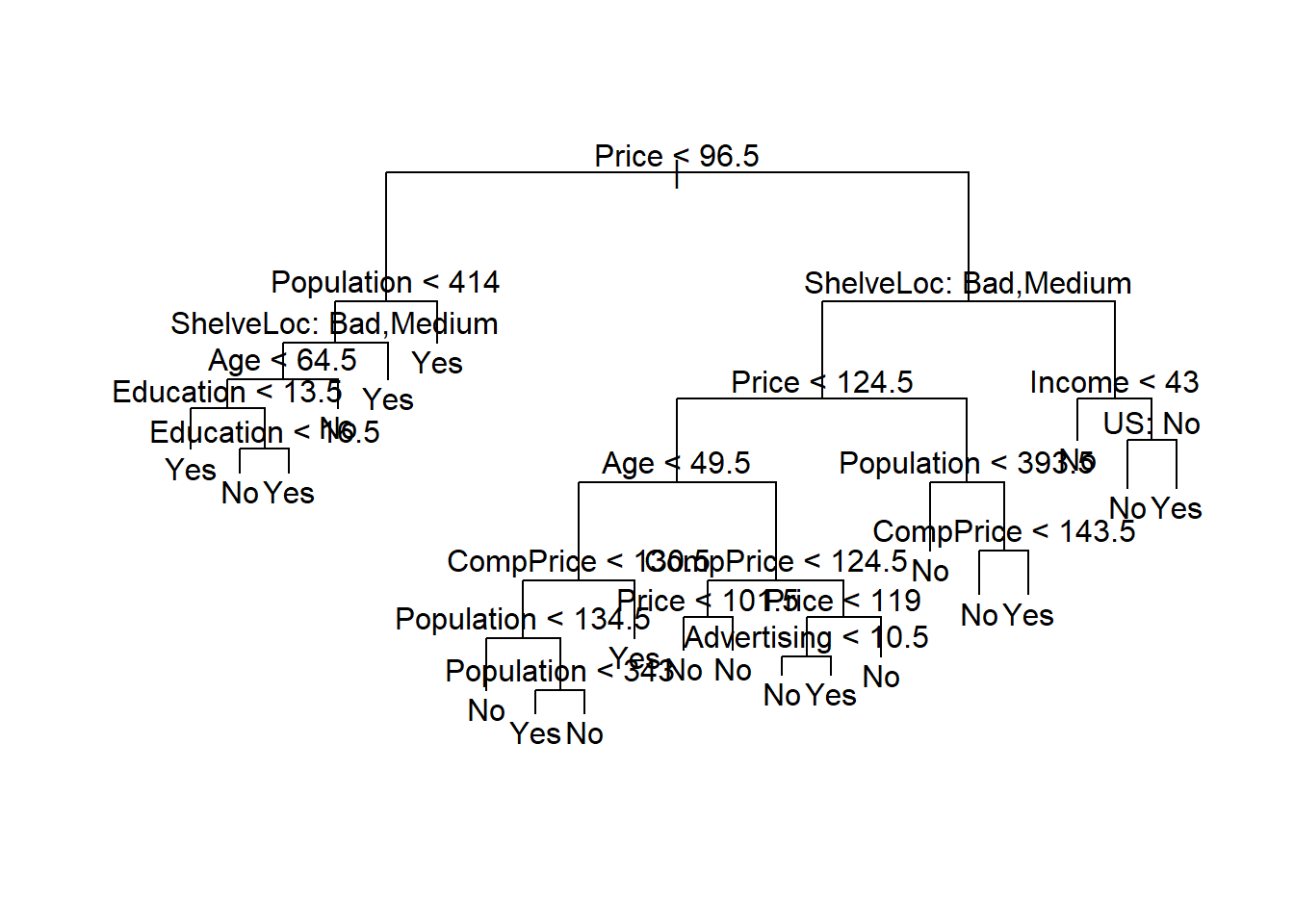
用剪枝后的树对测试集进行预测,计算误判率:
## test.high
## pred3 No Yes
## No 104 32
## Yes 13 51## [1] 0.22516.3.2 随机森林
对训练集用随机森林法:
##
## Call:
## randomForest(formula = High ~ . - Sales, data = d, importance = TRUE, subset = train)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 3
##
## OOB estimate of error rate: 27%
## Confusion matrix:
## No Yes class.error
## No 100 19 0.1596639
## Yes 35 46 0.4320988这里mtry取缺省值,对应于随机森林法。
对测试集进行预报:
## test.high
## pred4 No Yes
## No 108 23
## Yes 9 60## [1] 0.16注意错判率结果依赖于训练集和测试集的划分, 另行选择训练集与测试集可能会得到很不一样的错判率结果。
对全集用随机森林:
##
## Call:
## randomForest(formula = High ~ . - Sales, data = d, importance = TRUE)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 3
##
## OOB estimate of error rate: 19%
## Confusion matrix:
## No Yes class.error
## No 209 27 0.1144068
## Yes 49 115 0.2987805各变量的重要度数值及其图形:
## No Yes MeanDecreaseAccuracy MeanDecreaseGini
## CompPrice 11.033758100 7.7402932 13.33895983 21.229763
## Income 3.866551200 3.5223698 5.31053942 19.861721
## Advertising 10.907742775 16.4431361 18.85477287 24.265021
## Population -1.036771688 -2.6857178 -2.48427062 15.324194
## Price 33.443509343 27.9971005 40.05127916 43.779513
## ShelveLoc 34.255084236 34.3178518 43.13153318 30.636062
## Age 9.704435069 11.0226748 14.66681661 22.262051
## Education -0.009866974 -0.1131696 -0.18869317 10.058102
## Urban 0.687787338 -0.9368078 -0.06090853 2.094715
## US 2.929166411 5.1334006 6.60885378 3.436709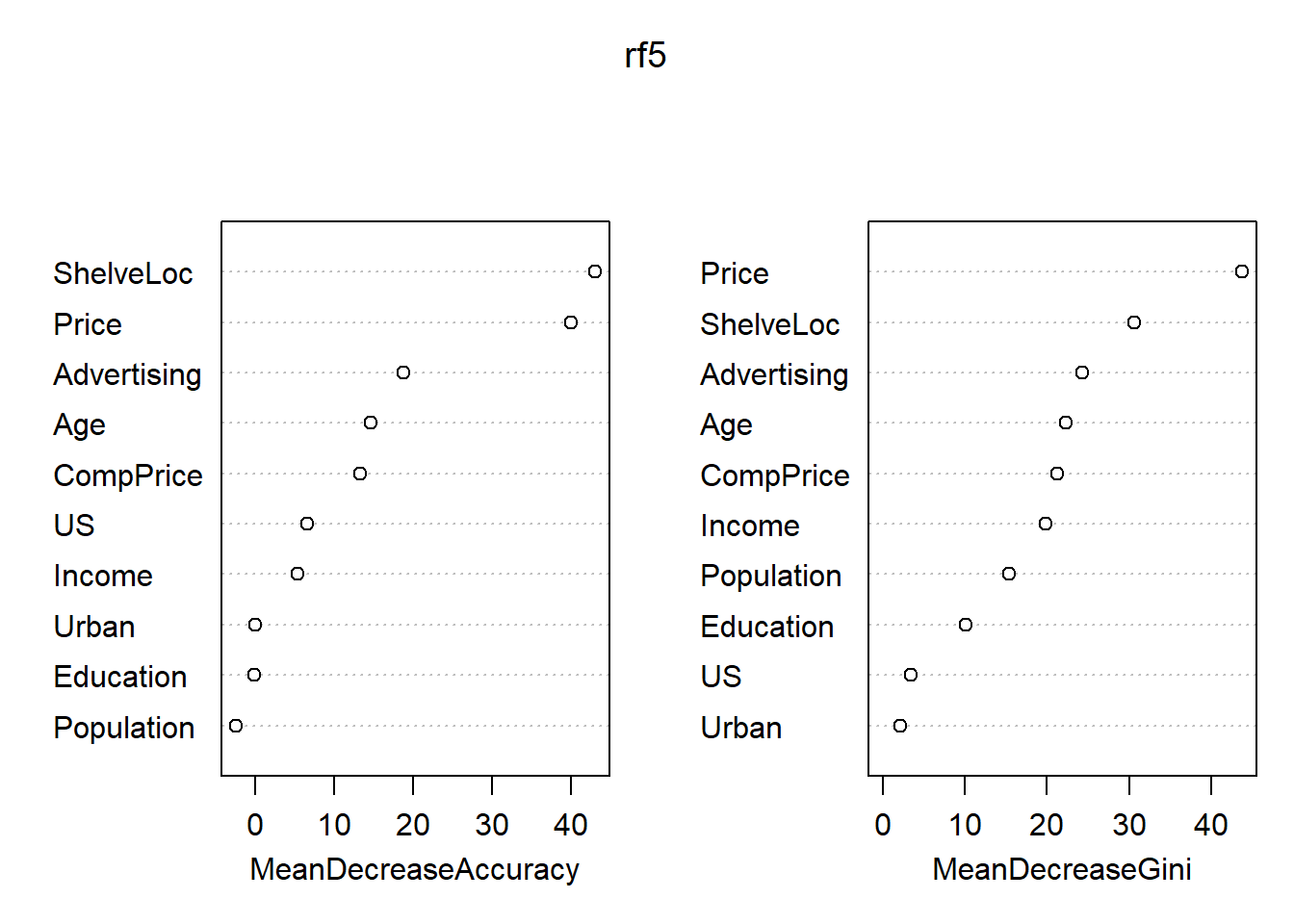
重要的自变量为Price, ShelfLoc, 其次有Age, Advertising, CompPrice, Income等。
16.4 提升法(Boosting)
提升法(Boosting)也是可以用在多种回归和判别问题中的方法。 提升法的想法是,用比较简单的模型拟合因变量, 计算残差, 然后以残差为新的因变量建模, 仍使用简单的模型, 把两次的回归函数作加权和, 得到新的残差后,再以新残差作为因变量建模, 如此重复地更新回归函数, 得到由多个回归函数加权和组成的最终的回归函数。
加权一般取为比较小的值, 其目的是降低逼近速度。 统计学习问题中降低逼近速度可以减轻过度拟合问题。
提升法算法:
(1)对训练集,设置\(r_i = y_i\),并令初始回归函数为\(\hat f(\cdot)=0\)。(2)对\(b=1,2,\dots,B\)重复执行:(a)以训练集的自变量为自变量,以\(r\)为因变量,拟合一个仅有\(d\)个分叉的简单树回归函数, 设为\(\hat f_b\);(b)更新回归函数,添加一个压缩过的树回归函数: \[\begin{aligned} \hat f(x) \leftarrow \hat f(x) + \lambda \hat f_b(x); \end{aligned}\](c)更新残差: \[\begin{aligned} r_i \leftarrow r_i - \lambda \hat f_b(x_i). \end{aligned}\]
(3)提升法的回归函数为 \[\begin{aligned} \hat f(x) = \sum_{b=1}^B \lambda \hat f_b(x) . \end{aligned}\]
用多少个回归函数做加权和,即\(B\)的选取问题。 取得\(B\)太大也会有过度拟合, 但是只要\(B\)不太大这个问题不严重。 可以用交叉验证选择\(B\)的值。
收缩系数\(\lambda\)是一个小的正数, 控制学习速度, 经常用0.01, 0.001这样的值, 与要解决的问题有关。 取\(\lambda\)很小,就需要取\(B\)很大。
用来控制每个回归函数复杂度的参数, 对树回归而言就是树的大小。 一个分叉的树往往就很好。 取单个分叉时结果模型是可加模型, 没有交互项, 这是因为每个加权相加得回归函数都只依赖于单一自变量。 \(d>1\)时就加入了交互项。
16.5 统计学习算法调用统一界面
不同的统计学习算法及其实现函数使用了不同的程序语法。 但是, 这些方法一般也有流程上的共同之处。 一般都将数据分为训练集与测试集, 在训练集上一般使用交叉验证方法求得调节参数的最优值, 然后在测试集上进行预测, 得到客观的模型效果评价。
R扩展包caret和mlr可以整合现有的统计学习函数, 用一个统一的调用界面访问统一的常用功能, 并可以实现交叉验证等功能。
16.6 波士顿郊区房价数据分析演示
MASS包的Boston数据包含了波士顿地区郊区房价的若干数据。以众位房价medv为因变量建立回归模型。 首先把缺失值去掉后存入数据集d:
数据集概况:
## 'data.frame': 506 obs. of 14 variables:
## $ crim : num 0.00632 0.02731 0.02729 0.03237 0.06905 ...
## $ zn : num 18 0 0 0 0 0 12.5 12.5 12.5 12.5 ...
## $ indus : num 2.31 7.07 7.07 2.18 2.18 2.18 7.87 7.87 7.87 7.87 ...
## $ chas : int 0 0 0 0 0 0 0 0 0 0 ...
## $ nox : num 0.538 0.469 0.469 0.458 0.458 0.458 0.524 0.524 0.524 0.524 ...
## $ rm : num 6.58 6.42 7.18 7 7.15 ...
## $ age : num 65.2 78.9 61.1 45.8 54.2 58.7 66.6 96.1 100 85.9 ...
## $ dis : num 4.09 4.97 4.97 6.06 6.06 ...
## $ rad : int 1 2 2 3 3 3 5 5 5 5 ...
## $ tax : num 296 242 242 222 222 222 311 311 311 311 ...
## $ ptratio: num 15.3 17.8 17.8 18.7 18.7 18.7 15.2 15.2 15.2 15.2 ...
## $ black : num 397 397 393 395 397 ...
## $ lstat : num 4.98 9.14 4.03 2.94 5.33 ...
## $ medv : num 24 21.6 34.7 33.4 36.2 28.7 22.9 27.1 16.5 18.9 ...## crim zn indus chas nox rm age dis rad tax ptratio black lstat medv
## Min. : 0.00632 Min. : 0.00 Min. : 0.46 Min. :0.00000 Min. :0.3850 Min. :3.561 Min. : 2.90 Min. : 1.130 Min. : 1.000 Min. :187.0 Min. :12.60 Min. : 0.32 Min. : 1.73 Min. : 5.00
## 1st Qu.: 0.08205 1st Qu.: 0.00 1st Qu.: 5.19 1st Qu.:0.00000 1st Qu.:0.4490 1st Qu.:5.886 1st Qu.: 45.02 1st Qu.: 2.100 1st Qu.: 4.000 1st Qu.:279.0 1st Qu.:17.40 1st Qu.:375.38 1st Qu.: 6.95 1st Qu.:17.02
## Median : 0.25651 Median : 0.00 Median : 9.69 Median :0.00000 Median :0.5380 Median :6.208 Median : 77.50 Median : 3.207 Median : 5.000 Median :330.0 Median :19.05 Median :391.44 Median :11.36 Median :21.20
## Mean : 3.61352 Mean : 11.36 Mean :11.14 Mean :0.06917 Mean :0.5547 Mean :6.285 Mean : 68.57 Mean : 3.795 Mean : 9.549 Mean :408.2 Mean :18.46 Mean :356.67 Mean :12.65 Mean :22.53
## 3rd Qu.: 3.67708 3rd Qu.: 12.50 3rd Qu.:18.10 3rd Qu.:0.00000 3rd Qu.:0.6240 3rd Qu.:6.623 3rd Qu.: 94.08 3rd Qu.: 5.188 3rd Qu.:24.000 3rd Qu.:666.0 3rd Qu.:20.20 3rd Qu.:396.23 3rd Qu.:16.95 3rd Qu.:25.00
## Max. :88.97620 Max. :100.00 Max. :27.74 Max. :1.00000 Max. :0.8710 Max. :8.780 Max. :100.00 Max. :12.127 Max. :24.000 Max. :711.0 Max. :22.00 Max. :396.90 Max. :37.97 Max. :50.0016.6.1 回归树
16.6.1.1 划分训练集和测试集
set.seed(1)
d <- na.omit(Boston)
train_id <- sample(nrow(d), size=round(0.5*nrow(d)))
train <- rep(FALSE, nrow(d))
train[train_id] <- TRUE
test <- (!train)对训练集建立未剪枝的树:
##
## Regression tree:
## tree(formula = medv ~ ., data = d, subset = train)
## Variables actually used in tree construction:
## [1] "rm" "lstat" "crim" "age"
## Number of terminal nodes: 7
## Residual mean deviance: 10.38 = 2555 / 246
## Distribution of residuals:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -10.1800 -1.7770 -0.1775 0.0000 1.9230 16.5800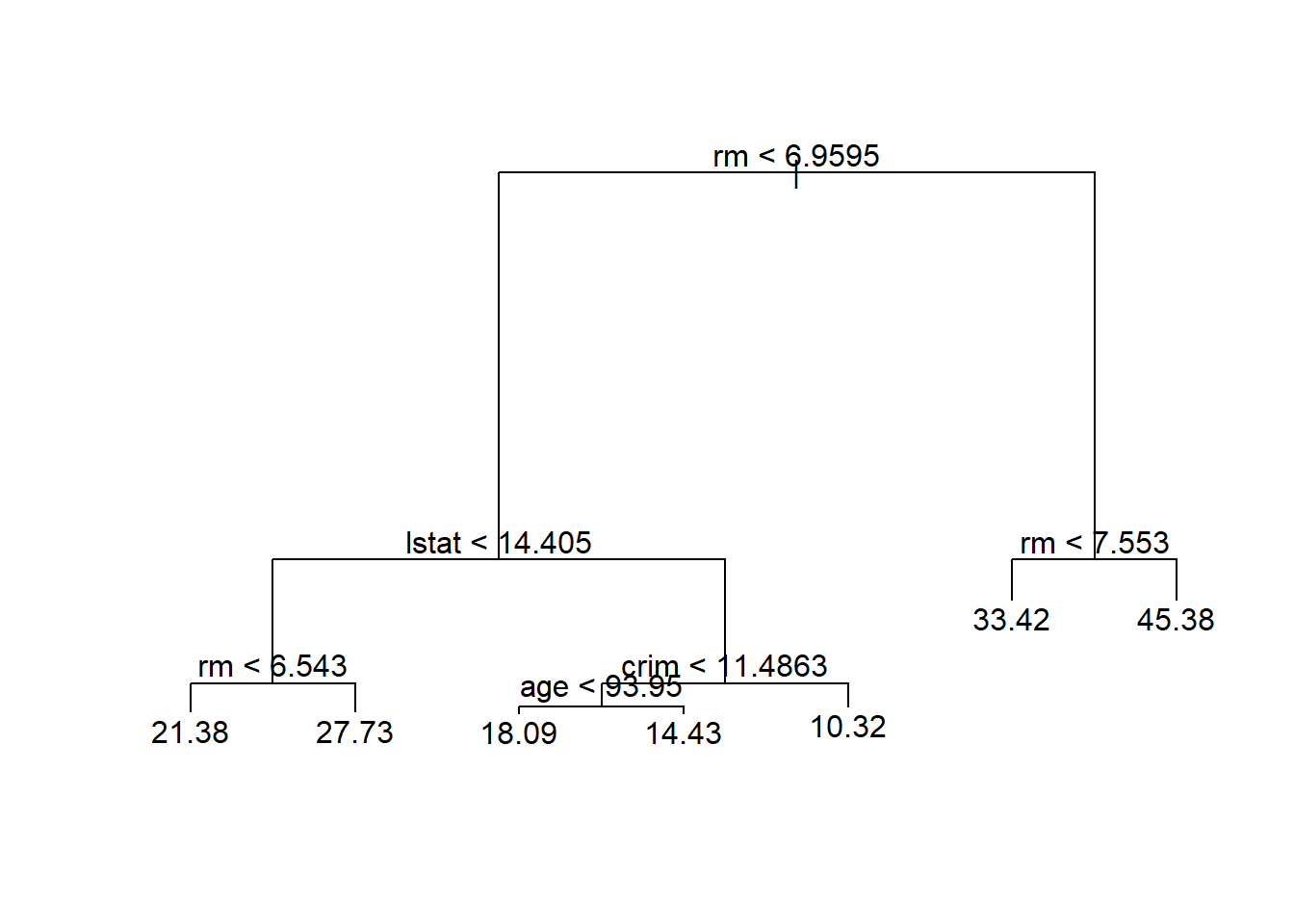
用未剪枝的树对测试集进行预测,计算均方误差:
## [1] 35.2868816.6.2 装袋法
用randomForest包计算。
当参数mtry取为自变量个数时按照装袋法计算。
对训练集计算。
set.seed(1)
bag1 <- randomForest(medv ~ ., data=d,
subset=train, mtry=ncol(d)-1, importance=TRUE)
bag1##
## Call:
## randomForest(formula = medv ~ ., data = d, mtry = ncol(d) - 1, importance = TRUE, subset = train)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 13
##
## Mean of squared residuals: 11.24028
## % Var explained: 85.38在测试集上计算装袋法的均方误差:
## [1] 23.2778比单棵树的结果有明显改善。
16.6.3 随机森林
用randomForest包计算。
当参数mtry取为缺省值时按照随机森林方法计算。
对训练集计算。
##
## Call:
## randomForest(formula = medv ~ ., data = d, importance = TRUE, subset = train)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 4
##
## Mean of squared residuals: 10.65524
## % Var explained: 86.14在测试集上计算随机森林法的均方误差:
## [1] 18.73938比单棵树的结果有明显改善, 比装袋法结果好。 但是,如果重新划分训练集合测试集, 结果可能就很不一样。
各变量的重要度数值及其图形:
## %IncMSE IncNodePurity
## crim 13.395947 1187.68244
## zn 3.764558 205.19223
## indus 6.634904 941.08254
## chas 2.556251 73.20277
## nox 13.848778 1092.57116
## rm 28.310083 6188.44612
## age 12.175066 659.38628
## dis 10.688898 958.36302
## rad 3.702112 151.38660
## tax 10.732676 516.77833
## ptratio 11.114057 1296.12067
## black 8.047270 336.29628
## lstat 25.983897 5396.58419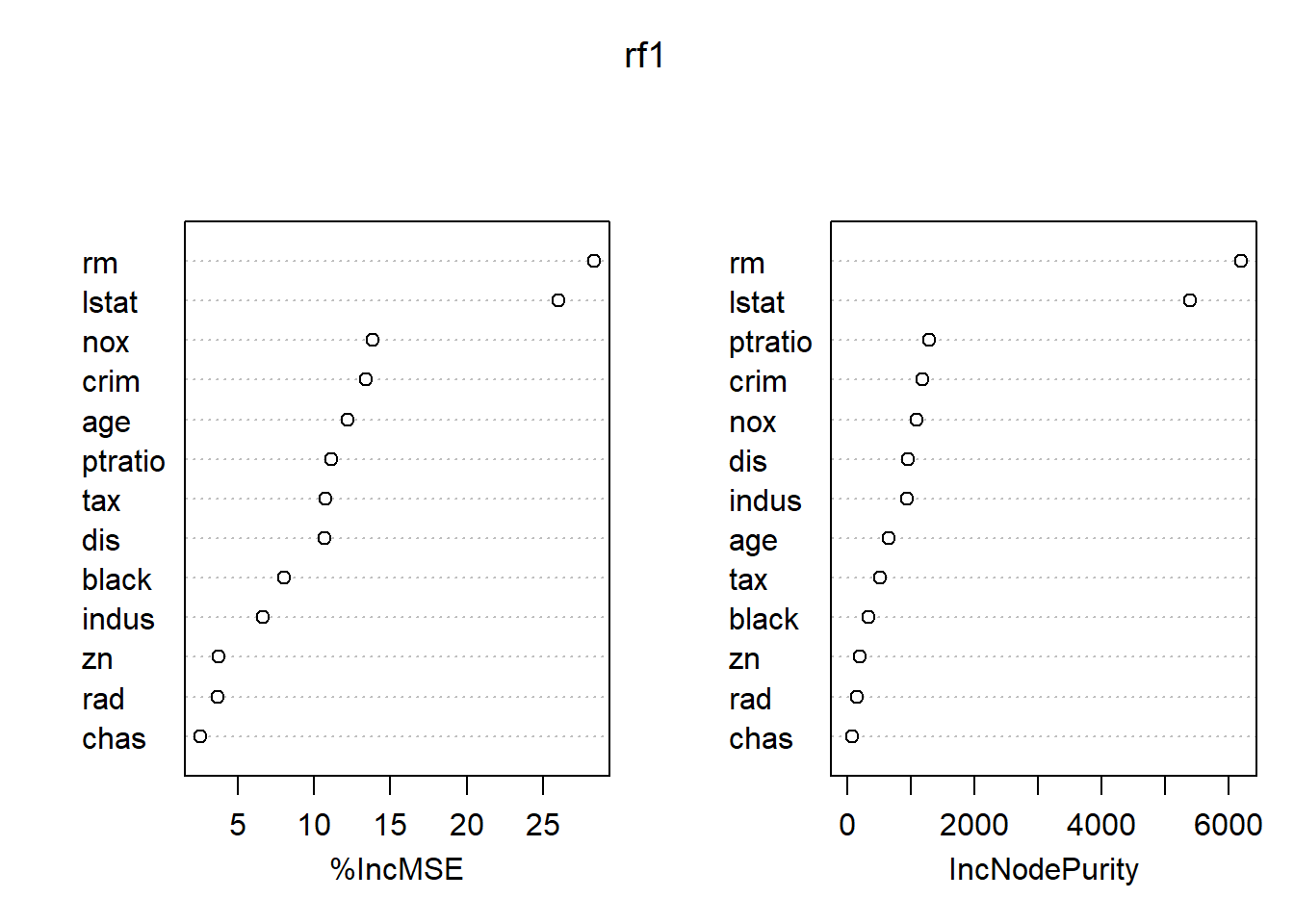
16.6.4 提升法
16.6.4.1 使用gbm包
这里使用gbm包。
在训练集上拟合:
set.seed(1)
bst1 <- gbm(
medv ~ .,
data=d[train,],
distribution='gaussian',
n.trees=5000,
interaction.depth=4)
summary(bst1)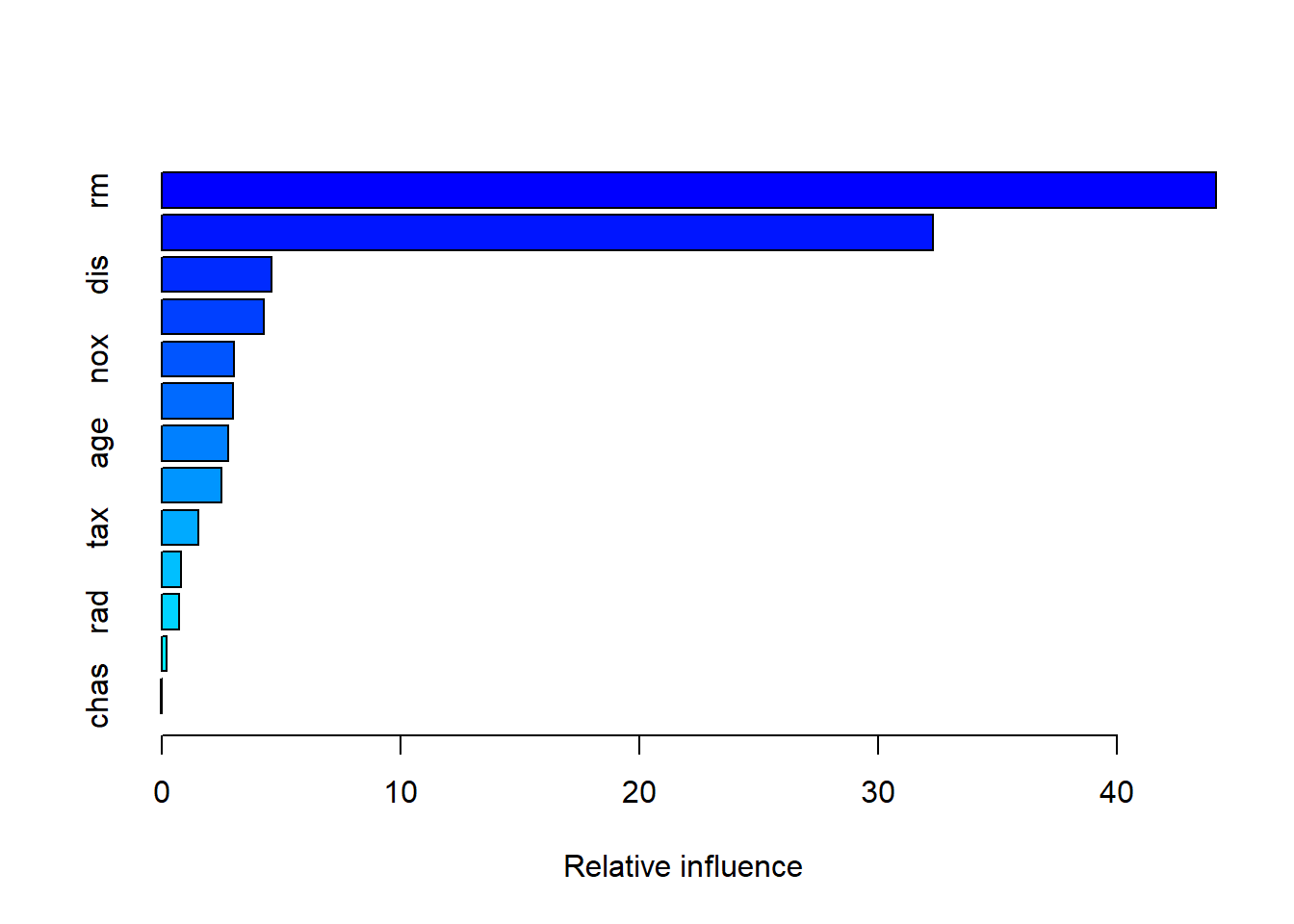
## var rel.inf
## rm rm 44.15951160
## lstat lstat 32.29267660
## dis dis 4.61448193
## crim crim 4.26335913
## nox nox 3.04501491
## black black 3.00055105
## age age 2.78125346
## ptratio ptratio 2.49234381
## tax tax 1.53209088
## indus indus 0.83074496
## rad rad 0.74565664
## zn zn 0.21988113
## chas chas 0.02243389lstat和rm是最重要的变量。
在测试集上预报,并计算均方误差:
## [1] 18.5659与随机森林方法结果相近。
如果提高学习速度:
bst2 <- gbm(
medv ~ .,
data=d[train,],
distribution='gaussian',
n.trees=5000,
interaction.depth=4,
shrinkage=0.2)
yhat <- predict(
bst2,
newdata=d[test,],
n.trees=5000)
mean( (yhat - d[test, 'medv'])^2 )## [1] 20.45657效果差不多。 但是,如果重新划分训练集和测试集, 结果可能很不一样。
16.6.4.2 使用xgboost包
使用xgboost计算。训练:
library(xgboost)
dx.train <- list(
data=as.matrix(d[train, -ncol(d)]),
label=d[["medv"]][train])
dx.test <- list(
data=as.matrix(d[test, -ncol(d)]),
label=d[["medv"]][test])
xg1 <- xgboost(
data = dx.train$data,
label = dx.train$label,
booster = "gbtree", # 基础模型
objective ="reg:squarederror", # 目标函数
max_depth = 2,
nrounds = 20) # 迭代次数## [1] train-rmse:16.550534
## [2] train-rmse:12.001012
## [3] train-rmse:8.828046
## [4] train-rmse:6.666827
## [5] train-rmse:5.179067
## [6] train-rmse:4.194855
## [7] train-rmse:3.574384
## [8] train-rmse:3.186898
## [9] train-rmse:2.915877
## [10] train-rmse:2.750005
## [11] train-rmse:2.631165
## [12] train-rmse:2.553116
## [13] train-rmse:2.484000
## [14] train-rmse:2.441754
## [15] train-rmse:2.374598
## [16] train-rmse:2.297728
## [17] train-rmse:2.239265
## [18] train-rmse:2.187598
## [19] train-rmse:2.161275
## [20] train-rmse:2.134788预测:
## [1] 20.28769比gbm结果差一些。
注意,xgboost的自变量需要是矩阵类型,
不支持数据框。
Boston数据集的自变量都是数值型的。
如果有数据中有因子型,
应该用Matrix::sparse.model.matrix()函数将数据框转换为数值型矩阵,
结果中的因子型变量被转换成哑变量形式。
16.6.4.3 使用lightgbm包
使用lightgbm计算。训练:
## Loading required package: R6##
## Attaching package: 'lightgbm'## The following objects are masked from 'package:xgboost':
##
## getinfo, setinfo, slicedl.train <- list(
data=as.matrix(d[train, -ncol(d)]),
label=d[["medv"]][train])
dl.test <- list(
data=as.matrix(d[test, -ncol(d)]),
label=d[["medv"]][test])
lgbm1 <- lightgbm(
data = dl.train$data,
label = dl.train$label,
obj ="regression", # 目标函数
max_depth = 2, # 树的深度
nrounds = 50) # 迭代次数## Warning in (function (params = list(), data, nrounds = 100L, valids = list(), : lgb.train: Found the following passed through '...': max_depth. These will be used, but in future releases of lightgbm, this warning will become an error. Add these to 'params' instead. See ?lgb.train for documentation on how to call this function.## [LightGBM] [Warning] Auto-choosing col-wise multi-threading, the overhead of testing was 0.000872 seconds.
## You can set `force_col_wise=true` to remove the overhead.
## [LightGBM] [Info] Total Bins 658
## [LightGBM] [Info] Number of data points in the train set: 253, number of used features: 12
## [LightGBM] [Info] Start training from score 21.786561
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[1]: train's l2:66.2431"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[2]: train's l2:57.636"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[3]: train's l2:50.5392"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[4]: train's l2:44.7832"
## [1] "[5]: train's l2:39.0265"
## [1] "[6]: train's l2:34.3006"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[7]: train's l2:30.6794"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[8]: train's l2:27.8866"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[9]: train's l2:25.2917"
## [1] "[10]: train's l2:22.7395"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[11]: train's l2:20.8562"
## [1] "[12]: train's l2:18.9931"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[13]: train's l2:17.6703"
## [1] "[14]: train's l2:16.2959"
## [1] "[15]: train's l2:15.1774"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[16]: train's l2:14.3231"
## [1] "[17]: train's l2:13.453"
## [1] "[18]: train's l2:12.7437"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[19]: train's l2:12.1731"
## [1] "[20]: train's l2:11.6418"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[21]: train's l2:11.2383"
## [1] "[22]: train's l2:10.7768"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[23]: train's l2:10.4449"
## [1] "[24]: train's l2:10.0823"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[25]: train's l2:9.84566"
## [1] "[26]: train's l2:9.56599"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[27]: train's l2:9.38185"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[28]: train's l2:9.21302"
## [1] "[29]: train's l2:8.96784"
## [1] "[30]: train's l2:8.76156"
## [1] "[31]: train's l2:8.56684"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[32]: train's l2:8.45506"
## [1] "[33]: train's l2:8.30198"
## [1] "[34]: train's l2:8.15626"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[35]: train's l2:8.0615"
## [1] "[36]: train's l2:7.91956"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[37]: train's l2:7.8353"
## [1] "[38]: train's l2:7.71917"
## [1] "[39]: train's l2:7.58138"
## [1] "[40]: train's l2:7.48035"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[41]: train's l2:7.40417"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[42]: train's l2:7.35067"
## [1] "[43]: train's l2:7.25805"
## [1] "[44]: train's l2:7.14817"
## [1] "[45]: train's l2:7.07001"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[46]: train's l2:7.0208"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[47]: train's l2:6.97039"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[48]: train's l2:6.90809"
## [1] "[49]: train's l2:6.84257"
## [LightGBM] [Warning] No further splits with positive gain, best gain: -inf
## [1] "[50]: train's l2:6.80303"预测:
## [1] 24.12621效果不好,误差比较大。 有可能需要调整参数。
注意,xgboost的自变量需要是矩阵类型,
不支持数据框。
Boston数据集的自变量都是数值型的。
如果有数据中有因子型,
应该用Matrix::sparse.model.matrix()函数将数据框转换为数值型矩阵,
结果中的因子型变量被转换成哑变量形式。
lightgbm对输入数据格式也有特殊要求, 详见文档。
16.7 附录:数据
16.7.1 Heart数据
| Age | Sex | ChestPain | RestBP | Chol | Fbs | RestECG | MaxHR | ExAng | Oldpeak | Slope | Ca | Thal | AHD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 63 | 1 | typical | 145 | 233 | 1 | 2 | 150 | 0 | 2.3 | 3 | 0 | fixed | No |
| 2 | 67 | 1 | asymptomatic | 160 | 286 | 0 | 2 | 108 | 1 | 1.5 | 2 | 3 | normal | Yes |
| 3 | 67 | 1 | asymptomatic | 120 | 229 | 0 | 2 | 129 | 1 | 2.6 | 2 | 2 | reversable | Yes |
| 4 | 37 | 1 | nonanginal | 130 | 250 | 0 | 0 | 187 | 0 | 3.5 | 3 | 0 | normal | No |
| 5 | 41 | 0 | nontypical | 130 | 204 | 0 | 2 | 172 | 0 | 1.4 | 1 | 0 | normal | No |
| 6 | 56 | 1 | nontypical | 120 | 236 | 0 | 0 | 178 | 0 | 0.8 | 1 | 0 | normal | No |
| 7 | 62 | 0 | asymptomatic | 140 | 268 | 0 | 2 | 160 | 0 | 3.6 | 3 | 2 | normal | Yes |
| 8 | 57 | 0 | asymptomatic | 120 | 354 | 0 | 0 | 163 | 1 | 0.6 | 1 | 0 | normal | No |
| 9 | 63 | 1 | asymptomatic | 130 | 254 | 0 | 2 | 147 | 0 | 1.4 | 2 | 1 | reversable | Yes |
| 10 | 53 | 1 | asymptomatic | 140 | 203 | 1 | 2 | 155 | 1 | 3.1 | 3 | 0 | reversable | Yes |
| 11 | 57 | 1 | asymptomatic | 140 | 192 | 0 | 0 | 148 | 0 | 0.4 | 2 | 0 | fixed | No |
| 12 | 56 | 0 | nontypical | 140 | 294 | 0 | 2 | 153 | 0 | 1.3 | 2 | 0 | normal | No |
| 13 | 56 | 1 | nonanginal | 130 | 256 | 1 | 2 | 142 | 1 | 0.6 | 2 | 1 | fixed | Yes |
| 14 | 44 | 1 | nontypical | 120 | 263 | 0 | 0 | 173 | 0 | 0.0 | 1 | 0 | reversable | No |
| 15 | 52 | 1 | nonanginal | 172 | 199 | 1 | 0 | 162 | 0 | 0.5 | 1 | 0 | reversable | No |
| 16 | 57 | 1 | nonanginal | 150 | 168 | 0 | 0 | 174 | 0 | 1.6 | 1 | 0 | normal | No |
| 17 | 48 | 1 | nontypical | 110 | 229 | 0 | 0 | 168 | 0 | 1.0 | 3 | 0 | reversable | Yes |
| 18 | 54 | 1 | asymptomatic | 140 | 239 | 0 | 0 | 160 | 0 | 1.2 | 1 | 0 | normal | No |
| 19 | 48 | 0 | nonanginal | 130 | 275 | 0 | 0 | 139 | 0 | 0.2 | 1 | 0 | normal | No |
| 20 | 49 | 1 | nontypical | 130 | 266 | 0 | 0 | 171 | 0 | 0.6 | 1 | 0 | normal | No |
| 21 | 64 | 1 | typical | 110 | 211 | 0 | 2 | 144 | 1 | 1.8 | 2 | 0 | normal | No |
| 22 | 58 | 0 | typical | 150 | 283 | 1 | 2 | 162 | 0 | 1.0 | 1 | 0 | normal | No |
| 23 | 58 | 1 | nontypical | 120 | 284 | 0 | 2 | 160 | 0 | 1.8 | 2 | 0 | normal | Yes |
| 24 | 58 | 1 | nonanginal | 132 | 224 | 0 | 2 | 173 | 0 | 3.2 | 1 | 2 | reversable | Yes |
| 25 | 60 | 1 | asymptomatic | 130 | 206 | 0 | 2 | 132 | 1 | 2.4 | 2 | 2 | reversable | Yes |
| 26 | 50 | 0 | nonanginal | 120 | 219 | 0 | 0 | 158 | 0 | 1.6 | 2 | 0 | normal | No |
| 27 | 58 | 0 | nonanginal | 120 | 340 | 0 | 0 | 172 | 0 | 0.0 | 1 | 0 | normal | No |
| 28 | 66 | 0 | typical | 150 | 226 | 0 | 0 | 114 | 0 | 2.6 | 3 | 0 | normal | No |
| 29 | 43 | 1 | asymptomatic | 150 | 247 | 0 | 0 | 171 | 0 | 1.5 | 1 | 0 | normal | No |
| 30 | 40 | 1 | asymptomatic | 110 | 167 | 0 | 2 | 114 | 1 | 2.0 | 2 | 0 | reversable | Yes |
| 31 | 69 | 0 | typical | 140 | 239 | 0 | 0 | 151 | 0 | 1.8 | 1 | 2 | normal | No |
| 32 | 60 | 1 | asymptomatic | 117 | 230 | 1 | 0 | 160 | 1 | 1.4 | 1 | 2 | reversable | Yes |
| 33 | 64 | 1 | nonanginal | 140 | 335 | 0 | 0 | 158 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 34 | 59 | 1 | asymptomatic | 135 | 234 | 0 | 0 | 161 | 0 | 0.5 | 2 | 0 | reversable | No |
| 35 | 44 | 1 | nonanginal | 130 | 233 | 0 | 0 | 179 | 1 | 0.4 | 1 | 0 | normal | No |
| 36 | 42 | 1 | asymptomatic | 140 | 226 | 0 | 0 | 178 | 0 | 0.0 | 1 | 0 | normal | No |
| 37 | 43 | 1 | asymptomatic | 120 | 177 | 0 | 2 | 120 | 1 | 2.5 | 2 | 0 | reversable | Yes |
| 38 | 57 | 1 | asymptomatic | 150 | 276 | 0 | 2 | 112 | 1 | 0.6 | 2 | 1 | fixed | Yes |
| 39 | 55 | 1 | asymptomatic | 132 | 353 | 0 | 0 | 132 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 40 | 61 | 1 | nonanginal | 150 | 243 | 1 | 0 | 137 | 1 | 1.0 | 2 | 0 | normal | No |
| 41 | 65 | 0 | asymptomatic | 150 | 225 | 0 | 2 | 114 | 0 | 1.0 | 2 | 3 | reversable | Yes |
| 42 | 40 | 1 | typical | 140 | 199 | 0 | 0 | 178 | 1 | 1.4 | 1 | 0 | reversable | No |
| 43 | 71 | 0 | nontypical | 160 | 302 | 0 | 0 | 162 | 0 | 0.4 | 1 | 2 | normal | No |
| 44 | 59 | 1 | nonanginal | 150 | 212 | 1 | 0 | 157 | 0 | 1.6 | 1 | 0 | normal | No |
| 45 | 61 | 0 | asymptomatic | 130 | 330 | 0 | 2 | 169 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 46 | 58 | 1 | nonanginal | 112 | 230 | 0 | 2 | 165 | 0 | 2.5 | 2 | 1 | reversable | Yes |
| 47 | 51 | 1 | nonanginal | 110 | 175 | 0 | 0 | 123 | 0 | 0.6 | 1 | 0 | normal | No |
| 48 | 50 | 1 | asymptomatic | 150 | 243 | 0 | 2 | 128 | 0 | 2.6 | 2 | 0 | reversable | Yes |
| 49 | 65 | 0 | nonanginal | 140 | 417 | 1 | 2 | 157 | 0 | 0.8 | 1 | 1 | normal | No |
| 50 | 53 | 1 | nonanginal | 130 | 197 | 1 | 2 | 152 | 0 | 1.2 | 3 | 0 | normal | No |
| 51 | 41 | 0 | nontypical | 105 | 198 | 0 | 0 | 168 | 0 | 0.0 | 1 | 1 | normal | No |
| 52 | 65 | 1 | asymptomatic | 120 | 177 | 0 | 0 | 140 | 0 | 0.4 | 1 | 0 | reversable | No |
| 53 | 44 | 1 | asymptomatic | 112 | 290 | 0 | 2 | 153 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 54 | 44 | 1 | nontypical | 130 | 219 | 0 | 2 | 188 | 0 | 0.0 | 1 | 0 | normal | No |
| 55 | 60 | 1 | asymptomatic | 130 | 253 | 0 | 0 | 144 | 1 | 1.4 | 1 | 1 | reversable | Yes |
| 56 | 54 | 1 | asymptomatic | 124 | 266 | 0 | 2 | 109 | 1 | 2.2 | 2 | 1 | reversable | Yes |
| 57 | 50 | 1 | nonanginal | 140 | 233 | 0 | 0 | 163 | 0 | 0.6 | 2 | 1 | reversable | Yes |
| 58 | 41 | 1 | asymptomatic | 110 | 172 | 0 | 2 | 158 | 0 | 0.0 | 1 | 0 | reversable | Yes |
| 59 | 54 | 1 | nonanginal | 125 | 273 | 0 | 2 | 152 | 0 | 0.5 | 3 | 1 | normal | No |
| 60 | 51 | 1 | typical | 125 | 213 | 0 | 2 | 125 | 1 | 1.4 | 1 | 1 | normal | No |
| 61 | 51 | 0 | asymptomatic | 130 | 305 | 0 | 0 | 142 | 1 | 1.2 | 2 | 0 | reversable | Yes |
| 62 | 46 | 0 | nonanginal | 142 | 177 | 0 | 2 | 160 | 1 | 1.4 | 3 | 0 | normal | No |
| 63 | 58 | 1 | asymptomatic | 128 | 216 | 0 | 2 | 131 | 1 | 2.2 | 2 | 3 | reversable | Yes |
| 64 | 54 | 0 | nonanginal | 135 | 304 | 1 | 0 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 65 | 54 | 1 | asymptomatic | 120 | 188 | 0 | 0 | 113 | 0 | 1.4 | 2 | 1 | reversable | Yes |
| 66 | 60 | 1 | asymptomatic | 145 | 282 | 0 | 2 | 142 | 1 | 2.8 | 2 | 2 | reversable | Yes |
| 67 | 60 | 1 | nonanginal | 140 | 185 | 0 | 2 | 155 | 0 | 3.0 | 2 | 0 | normal | Yes |
| 68 | 54 | 1 | nonanginal | 150 | 232 | 0 | 2 | 165 | 0 | 1.6 | 1 | 0 | reversable | No |
| 69 | 59 | 1 | asymptomatic | 170 | 326 | 0 | 2 | 140 | 1 | 3.4 | 3 | 0 | reversable | Yes |
| 70 | 46 | 1 | nonanginal | 150 | 231 | 0 | 0 | 147 | 0 | 3.6 | 2 | 0 | normal | Yes |
| 71 | 65 | 0 | nonanginal | 155 | 269 | 0 | 0 | 148 | 0 | 0.8 | 1 | 0 | normal | No |
| 72 | 67 | 1 | asymptomatic | 125 | 254 | 1 | 0 | 163 | 0 | 0.2 | 2 | 2 | reversable | Yes |
| 73 | 62 | 1 | asymptomatic | 120 | 267 | 0 | 0 | 99 | 1 | 1.8 | 2 | 2 | reversable | Yes |
| 74 | 65 | 1 | asymptomatic | 110 | 248 | 0 | 2 | 158 | 0 | 0.6 | 1 | 2 | fixed | Yes |
| 75 | 44 | 1 | asymptomatic | 110 | 197 | 0 | 2 | 177 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 76 | 65 | 0 | nonanginal | 160 | 360 | 0 | 2 | 151 | 0 | 0.8 | 1 | 0 | normal | No |
| 77 | 60 | 1 | asymptomatic | 125 | 258 | 0 | 2 | 141 | 1 | 2.8 | 2 | 1 | reversable | Yes |
| 78 | 51 | 0 | nonanginal | 140 | 308 | 0 | 2 | 142 | 0 | 1.5 | 1 | 1 | normal | No |
| 79 | 48 | 1 | nontypical | 130 | 245 | 0 | 2 | 180 | 0 | 0.2 | 2 | 0 | normal | No |
| 80 | 58 | 1 | asymptomatic | 150 | 270 | 0 | 2 | 111 | 1 | 0.8 | 1 | 0 | reversable | Yes |
| 81 | 45 | 1 | asymptomatic | 104 | 208 | 0 | 2 | 148 | 1 | 3.0 | 2 | 0 | normal | No |
| 82 | 53 | 0 | asymptomatic | 130 | 264 | 0 | 2 | 143 | 0 | 0.4 | 2 | 0 | normal | No |
| 83 | 39 | 1 | nonanginal | 140 | 321 | 0 | 2 | 182 | 0 | 0.0 | 1 | 0 | normal | No |
| 84 | 68 | 1 | nonanginal | 180 | 274 | 1 | 2 | 150 | 1 | 1.6 | 2 | 0 | reversable | Yes |
| 85 | 52 | 1 | nontypical | 120 | 325 | 0 | 0 | 172 | 0 | 0.2 | 1 | 0 | normal | No |
| 86 | 44 | 1 | nonanginal | 140 | 235 | 0 | 2 | 180 | 0 | 0.0 | 1 | 0 | normal | No |
| 87 | 47 | 1 | nonanginal | 138 | 257 | 0 | 2 | 156 | 0 | 0.0 | 1 | 0 | normal | No |
| 89 | 53 | 0 | asymptomatic | 138 | 234 | 0 | 2 | 160 | 0 | 0.0 | 1 | 0 | normal | No |
| 90 | 51 | 0 | nonanginal | 130 | 256 | 0 | 2 | 149 | 0 | 0.5 | 1 | 0 | normal | No |
| 91 | 66 | 1 | asymptomatic | 120 | 302 | 0 | 2 | 151 | 0 | 0.4 | 2 | 0 | normal | No |
| 92 | 62 | 0 | asymptomatic | 160 | 164 | 0 | 2 | 145 | 0 | 6.2 | 3 | 3 | reversable | Yes |
| 93 | 62 | 1 | nonanginal | 130 | 231 | 0 | 0 | 146 | 0 | 1.8 | 2 | 3 | reversable | No |
| 94 | 44 | 0 | nonanginal | 108 | 141 | 0 | 0 | 175 | 0 | 0.6 | 2 | 0 | normal | No |
| 95 | 63 | 0 | nonanginal | 135 | 252 | 0 | 2 | 172 | 0 | 0.0 | 1 | 0 | normal | No |
| 96 | 52 | 1 | asymptomatic | 128 | 255 | 0 | 0 | 161 | 1 | 0.0 | 1 | 1 | reversable | Yes |
| 97 | 59 | 1 | asymptomatic | 110 | 239 | 0 | 2 | 142 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 98 | 60 | 0 | asymptomatic | 150 | 258 | 0 | 2 | 157 | 0 | 2.6 | 2 | 2 | reversable | Yes |
| 99 | 52 | 1 | nontypical | 134 | 201 | 0 | 0 | 158 | 0 | 0.8 | 1 | 1 | normal | No |
| 100 | 48 | 1 | asymptomatic | 122 | 222 | 0 | 2 | 186 | 0 | 0.0 | 1 | 0 | normal | No |
| 101 | 45 | 1 | asymptomatic | 115 | 260 | 0 | 2 | 185 | 0 | 0.0 | 1 | 0 | normal | No |
| 102 | 34 | 1 | typical | 118 | 182 | 0 | 2 | 174 | 0 | 0.0 | 1 | 0 | normal | No |
| 103 | 57 | 0 | asymptomatic | 128 | 303 | 0 | 2 | 159 | 0 | 0.0 | 1 | 1 | normal | No |
| 104 | 71 | 0 | nonanginal | 110 | 265 | 1 | 2 | 130 | 0 | 0.0 | 1 | 1 | normal | No |
| 105 | 49 | 1 | nonanginal | 120 | 188 | 0 | 0 | 139 | 0 | 2.0 | 2 | 3 | reversable | Yes |
| 106 | 54 | 1 | nontypical | 108 | 309 | 0 | 0 | 156 | 0 | 0.0 | 1 | 0 | reversable | No |
| 107 | 59 | 1 | asymptomatic | 140 | 177 | 0 | 0 | 162 | 1 | 0.0 | 1 | 1 | reversable | Yes |
| 108 | 57 | 1 | nonanginal | 128 | 229 | 0 | 2 | 150 | 0 | 0.4 | 2 | 1 | reversable | Yes |
| 109 | 61 | 1 | asymptomatic | 120 | 260 | 0 | 0 | 140 | 1 | 3.6 | 2 | 1 | reversable | Yes |
| 110 | 39 | 1 | asymptomatic | 118 | 219 | 0 | 0 | 140 | 0 | 1.2 | 2 | 0 | reversable | Yes |
| 111 | 61 | 0 | asymptomatic | 145 | 307 | 0 | 2 | 146 | 1 | 1.0 | 2 | 0 | reversable | Yes |
| 112 | 56 | 1 | asymptomatic | 125 | 249 | 1 | 2 | 144 | 1 | 1.2 | 2 | 1 | normal | Yes |
| 113 | 52 | 1 | typical | 118 | 186 | 0 | 2 | 190 | 0 | 0.0 | 2 | 0 | fixed | No |
| 114 | 43 | 0 | asymptomatic | 132 | 341 | 1 | 2 | 136 | 1 | 3.0 | 2 | 0 | reversable | Yes |
| 115 | 62 | 0 | nonanginal | 130 | 263 | 0 | 0 | 97 | 0 | 1.2 | 2 | 1 | reversable | Yes |
| 116 | 41 | 1 | nontypical | 135 | 203 | 0 | 0 | 132 | 0 | 0.0 | 2 | 0 | fixed | No |
| 117 | 58 | 1 | nonanginal | 140 | 211 | 1 | 2 | 165 | 0 | 0.0 | 1 | 0 | normal | No |
| 118 | 35 | 0 | asymptomatic | 138 | 183 | 0 | 0 | 182 | 0 | 1.4 | 1 | 0 | normal | No |
| 119 | 63 | 1 | asymptomatic | 130 | 330 | 1 | 2 | 132 | 1 | 1.8 | 1 | 3 | reversable | Yes |
| 120 | 65 | 1 | asymptomatic | 135 | 254 | 0 | 2 | 127 | 0 | 2.8 | 2 | 1 | reversable | Yes |
| 121 | 48 | 1 | asymptomatic | 130 | 256 | 1 | 2 | 150 | 1 | 0.0 | 1 | 2 | reversable | Yes |
| 122 | 63 | 0 | asymptomatic | 150 | 407 | 0 | 2 | 154 | 0 | 4.0 | 2 | 3 | reversable | Yes |
| 123 | 51 | 1 | nonanginal | 100 | 222 | 0 | 0 | 143 | 1 | 1.2 | 2 | 0 | normal | No |
| 124 | 55 | 1 | asymptomatic | 140 | 217 | 0 | 0 | 111 | 1 | 5.6 | 3 | 0 | reversable | Yes |
| 125 | 65 | 1 | typical | 138 | 282 | 1 | 2 | 174 | 0 | 1.4 | 2 | 1 | normal | Yes |
| 126 | 45 | 0 | nontypical | 130 | 234 | 0 | 2 | 175 | 0 | 0.6 | 2 | 0 | normal | No |
| 127 | 56 | 0 | asymptomatic | 200 | 288 | 1 | 2 | 133 | 1 | 4.0 | 3 | 2 | reversable | Yes |
| 128 | 54 | 1 | asymptomatic | 110 | 239 | 0 | 0 | 126 | 1 | 2.8 | 2 | 1 | reversable | Yes |
| 129 | 44 | 1 | nontypical | 120 | 220 | 0 | 0 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 130 | 62 | 0 | asymptomatic | 124 | 209 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 131 | 54 | 1 | nonanginal | 120 | 258 | 0 | 2 | 147 | 0 | 0.4 | 2 | 0 | reversable | No |
| 132 | 51 | 1 | nonanginal | 94 | 227 | 0 | 0 | 154 | 1 | 0.0 | 1 | 1 | reversable | No |
| 133 | 29 | 1 | nontypical | 130 | 204 | 0 | 2 | 202 | 0 | 0.0 | 1 | 0 | normal | No |
| 134 | 51 | 1 | asymptomatic | 140 | 261 | 0 | 2 | 186 | 1 | 0.0 | 1 | 0 | normal | No |
| 135 | 43 | 0 | nonanginal | 122 | 213 | 0 | 0 | 165 | 0 | 0.2 | 2 | 0 | normal | No |
| 136 | 55 | 0 | nontypical | 135 | 250 | 0 | 2 | 161 | 0 | 1.4 | 2 | 0 | normal | No |
| 137 | 70 | 1 | asymptomatic | 145 | 174 | 0 | 0 | 125 | 1 | 2.6 | 3 | 0 | reversable | Yes |
| 138 | 62 | 1 | nontypical | 120 | 281 | 0 | 2 | 103 | 0 | 1.4 | 2 | 1 | reversable | Yes |
| 139 | 35 | 1 | asymptomatic | 120 | 198 | 0 | 0 | 130 | 1 | 1.6 | 2 | 0 | reversable | Yes |
| 140 | 51 | 1 | nonanginal | 125 | 245 | 1 | 2 | 166 | 0 | 2.4 | 2 | 0 | normal | No |
| 141 | 59 | 1 | nontypical | 140 | 221 | 0 | 0 | 164 | 1 | 0.0 | 1 | 0 | normal | No |
| 142 | 59 | 1 | typical | 170 | 288 | 0 | 2 | 159 | 0 | 0.2 | 2 | 0 | reversable | Yes |
| 143 | 52 | 1 | nontypical | 128 | 205 | 1 | 0 | 184 | 0 | 0.0 | 1 | 0 | normal | No |
| 144 | 64 | 1 | nonanginal | 125 | 309 | 0 | 0 | 131 | 1 | 1.8 | 2 | 0 | reversable | Yes |
| 145 | 58 | 1 | nonanginal | 105 | 240 | 0 | 2 | 154 | 1 | 0.6 | 2 | 0 | reversable | No |
| 146 | 47 | 1 | nonanginal | 108 | 243 | 0 | 0 | 152 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 147 | 57 | 1 | asymptomatic | 165 | 289 | 1 | 2 | 124 | 0 | 1.0 | 2 | 3 | reversable | Yes |
| 148 | 41 | 1 | nonanginal | 112 | 250 | 0 | 0 | 179 | 0 | 0.0 | 1 | 0 | normal | No |
| 149 | 45 | 1 | nontypical | 128 | 308 | 0 | 2 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 150 | 60 | 0 | nonanginal | 102 | 318 | 0 | 0 | 160 | 0 | 0.0 | 1 | 1 | normal | No |
| 151 | 52 | 1 | typical | 152 | 298 | 1 | 0 | 178 | 0 | 1.2 | 2 | 0 | reversable | No |
| 152 | 42 | 0 | asymptomatic | 102 | 265 | 0 | 2 | 122 | 0 | 0.6 | 2 | 0 | normal | No |
| 153 | 67 | 0 | nonanginal | 115 | 564 | 0 | 2 | 160 | 0 | 1.6 | 2 | 0 | reversable | No |
| 154 | 55 | 1 | asymptomatic | 160 | 289 | 0 | 2 | 145 | 1 | 0.8 | 2 | 1 | reversable | Yes |
| 155 | 64 | 1 | asymptomatic | 120 | 246 | 0 | 2 | 96 | 1 | 2.2 | 3 | 1 | normal | Yes |
| 156 | 70 | 1 | asymptomatic | 130 | 322 | 0 | 2 | 109 | 0 | 2.4 | 2 | 3 | normal | Yes |
| 157 | 51 | 1 | asymptomatic | 140 | 299 | 0 | 0 | 173 | 1 | 1.6 | 1 | 0 | reversable | Yes |
| 158 | 58 | 1 | asymptomatic | 125 | 300 | 0 | 2 | 171 | 0 | 0.0 | 1 | 2 | reversable | Yes |
| 159 | 60 | 1 | asymptomatic | 140 | 293 | 0 | 2 | 170 | 0 | 1.2 | 2 | 2 | reversable | Yes |
| 160 | 68 | 1 | nonanginal | 118 | 277 | 0 | 0 | 151 | 0 | 1.0 | 1 | 1 | reversable | No |
| 161 | 46 | 1 | nontypical | 101 | 197 | 1 | 0 | 156 | 0 | 0.0 | 1 | 0 | reversable | No |
| 162 | 77 | 1 | asymptomatic | 125 | 304 | 0 | 2 | 162 | 1 | 0.0 | 1 | 3 | normal | Yes |
| 163 | 54 | 0 | nonanginal | 110 | 214 | 0 | 0 | 158 | 0 | 1.6 | 2 | 0 | normal | No |
| 164 | 58 | 0 | asymptomatic | 100 | 248 | 0 | 2 | 122 | 0 | 1.0 | 2 | 0 | normal | No |
| 165 | 48 | 1 | nonanginal | 124 | 255 | 1 | 0 | 175 | 0 | 0.0 | 1 | 2 | normal | No |
| 166 | 57 | 1 | asymptomatic | 132 | 207 | 0 | 0 | 168 | 1 | 0.0 | 1 | 0 | reversable | No |
| 168 | 54 | 0 | nontypical | 132 | 288 | 1 | 2 | 159 | 1 | 0.0 | 1 | 1 | normal | No |
| 169 | 35 | 1 | asymptomatic | 126 | 282 | 0 | 2 | 156 | 1 | 0.0 | 1 | 0 | reversable | Yes |
| 170 | 45 | 0 | nontypical | 112 | 160 | 0 | 0 | 138 | 0 | 0.0 | 2 | 0 | normal | No |
| 171 | 70 | 1 | nonanginal | 160 | 269 | 0 | 0 | 112 | 1 | 2.9 | 2 | 1 | reversable | Yes |
| 172 | 53 | 1 | asymptomatic | 142 | 226 | 0 | 2 | 111 | 1 | 0.0 | 1 | 0 | reversable | No |
| 173 | 59 | 0 | asymptomatic | 174 | 249 | 0 | 0 | 143 | 1 | 0.0 | 2 | 0 | normal | Yes |
| 174 | 62 | 0 | asymptomatic | 140 | 394 | 0 | 2 | 157 | 0 | 1.2 | 2 | 0 | normal | No |
| 175 | 64 | 1 | asymptomatic | 145 | 212 | 0 | 2 | 132 | 0 | 2.0 | 2 | 2 | fixed | Yes |
| 176 | 57 | 1 | asymptomatic | 152 | 274 | 0 | 0 | 88 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 177 | 52 | 1 | asymptomatic | 108 | 233 | 1 | 0 | 147 | 0 | 0.1 | 1 | 3 | reversable | No |
| 178 | 56 | 1 | asymptomatic | 132 | 184 | 0 | 2 | 105 | 1 | 2.1 | 2 | 1 | fixed | Yes |
| 179 | 43 | 1 | nonanginal | 130 | 315 | 0 | 0 | 162 | 0 | 1.9 | 1 | 1 | normal | No |
| 180 | 53 | 1 | nonanginal | 130 | 246 | 1 | 2 | 173 | 0 | 0.0 | 1 | 3 | normal | No |
| 181 | 48 | 1 | asymptomatic | 124 | 274 | 0 | 2 | 166 | 0 | 0.5 | 2 | 0 | reversable | Yes |
| 182 | 56 | 0 | asymptomatic | 134 | 409 | 0 | 2 | 150 | 1 | 1.9 | 2 | 2 | reversable | Yes |
| 183 | 42 | 1 | typical | 148 | 244 | 0 | 2 | 178 | 0 | 0.8 | 1 | 2 | normal | No |
| 184 | 59 | 1 | typical | 178 | 270 | 0 | 2 | 145 | 0 | 4.2 | 3 | 0 | reversable | No |
| 185 | 60 | 0 | asymptomatic | 158 | 305 | 0 | 2 | 161 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 186 | 63 | 0 | nontypical | 140 | 195 | 0 | 0 | 179 | 0 | 0.0 | 1 | 2 | normal | No |
| 187 | 42 | 1 | nonanginal | 120 | 240 | 1 | 0 | 194 | 0 | 0.8 | 3 | 0 | reversable | No |
| 188 | 66 | 1 | nontypical | 160 | 246 | 0 | 0 | 120 | 1 | 0.0 | 2 | 3 | fixed | Yes |
| 189 | 54 | 1 | nontypical | 192 | 283 | 0 | 2 | 195 | 0 | 0.0 | 1 | 1 | reversable | Yes |
| 190 | 69 | 1 | nonanginal | 140 | 254 | 0 | 2 | 146 | 0 | 2.0 | 2 | 3 | reversable | Yes |
| 191 | 50 | 1 | nonanginal | 129 | 196 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 192 | 51 | 1 | asymptomatic | 140 | 298 | 0 | 0 | 122 | 1 | 4.2 | 2 | 3 | reversable | Yes |
| 194 | 62 | 0 | asymptomatic | 138 | 294 | 1 | 0 | 106 | 0 | 1.9 | 2 | 3 | normal | Yes |
| 195 | 68 | 0 | nonanginal | 120 | 211 | 0 | 2 | 115 | 0 | 1.5 | 2 | 0 | normal | No |
| 196 | 67 | 1 | asymptomatic | 100 | 299 | 0 | 2 | 125 | 1 | 0.9 | 2 | 2 | normal | Yes |
| 197 | 69 | 1 | typical | 160 | 234 | 1 | 2 | 131 | 0 | 0.1 | 2 | 1 | normal | No |
| 198 | 45 | 0 | asymptomatic | 138 | 236 | 0 | 2 | 152 | 1 | 0.2 | 2 | 0 | normal | No |
| 199 | 50 | 0 | nontypical | 120 | 244 | 0 | 0 | 162 | 0 | 1.1 | 1 | 0 | normal | No |
| 200 | 59 | 1 | typical | 160 | 273 | 0 | 2 | 125 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 201 | 50 | 0 | asymptomatic | 110 | 254 | 0 | 2 | 159 | 0 | 0.0 | 1 | 0 | normal | No |
| 202 | 64 | 0 | asymptomatic | 180 | 325 | 0 | 0 | 154 | 1 | 0.0 | 1 | 0 | normal | No |
| 203 | 57 | 1 | nonanginal | 150 | 126 | 1 | 0 | 173 | 0 | 0.2 | 1 | 1 | reversable | No |
| 204 | 64 | 0 | nonanginal | 140 | 313 | 0 | 0 | 133 | 0 | 0.2 | 1 | 0 | reversable | No |
| 205 | 43 | 1 | asymptomatic | 110 | 211 | 0 | 0 | 161 | 0 | 0.0 | 1 | 0 | reversable | No |
| 206 | 45 | 1 | asymptomatic | 142 | 309 | 0 | 2 | 147 | 1 | 0.0 | 2 | 3 | reversable | Yes |
| 207 | 58 | 1 | asymptomatic | 128 | 259 | 0 | 2 | 130 | 1 | 3.0 | 2 | 2 | reversable | Yes |
| 208 | 50 | 1 | asymptomatic | 144 | 200 | 0 | 2 | 126 | 1 | 0.9 | 2 | 0 | reversable | Yes |
| 209 | 55 | 1 | nontypical | 130 | 262 | 0 | 0 | 155 | 0 | 0.0 | 1 | 0 | normal | No |
| 210 | 62 | 0 | asymptomatic | 150 | 244 | 0 | 0 | 154 | 1 | 1.4 | 2 | 0 | normal | Yes |
| 211 | 37 | 0 | nonanginal | 120 | 215 | 0 | 0 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 212 | 38 | 1 | typical | 120 | 231 | 0 | 0 | 182 | 1 | 3.8 | 2 | 0 | reversable | Yes |
| 213 | 41 | 1 | nonanginal | 130 | 214 | 0 | 2 | 168 | 0 | 2.0 | 2 | 0 | normal | No |
| 214 | 66 | 0 | asymptomatic | 178 | 228 | 1 | 0 | 165 | 1 | 1.0 | 2 | 2 | reversable | Yes |
| 215 | 52 | 1 | asymptomatic | 112 | 230 | 0 | 0 | 160 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 216 | 56 | 1 | typical | 120 | 193 | 0 | 2 | 162 | 0 | 1.9 | 2 | 0 | reversable | No |
| 217 | 46 | 0 | nontypical | 105 | 204 | 0 | 0 | 172 | 0 | 0.0 | 1 | 0 | normal | No |
| 218 | 46 | 0 | asymptomatic | 138 | 243 | 0 | 2 | 152 | 1 | 0.0 | 2 | 0 | normal | No |
| 219 | 64 | 0 | asymptomatic | 130 | 303 | 0 | 0 | 122 | 0 | 2.0 | 2 | 2 | normal | No |
| 220 | 59 | 1 | asymptomatic | 138 | 271 | 0 | 2 | 182 | 0 | 0.0 | 1 | 0 | normal | No |
| 221 | 41 | 0 | nonanginal | 112 | 268 | 0 | 2 | 172 | 1 | 0.0 | 1 | 0 | normal | No |
| 222 | 54 | 0 | nonanginal | 108 | 267 | 0 | 2 | 167 | 0 | 0.0 | 1 | 0 | normal | No |
| 223 | 39 | 0 | nonanginal | 94 | 199 | 0 | 0 | 179 | 0 | 0.0 | 1 | 0 | normal | No |
| 224 | 53 | 1 | asymptomatic | 123 | 282 | 0 | 0 | 95 | 1 | 2.0 | 2 | 2 | reversable | Yes |
| 225 | 63 | 0 | asymptomatic | 108 | 269 | 0 | 0 | 169 | 1 | 1.8 | 2 | 2 | normal | Yes |
| 226 | 34 | 0 | nontypical | 118 | 210 | 0 | 0 | 192 | 0 | 0.7 | 1 | 0 | normal | No |
| 227 | 47 | 1 | asymptomatic | 112 | 204 | 0 | 0 | 143 | 0 | 0.1 | 1 | 0 | normal | No |
| 228 | 67 | 0 | nonanginal | 152 | 277 | 0 | 0 | 172 | 0 | 0.0 | 1 | 1 | normal | No |
| 229 | 54 | 1 | asymptomatic | 110 | 206 | 0 | 2 | 108 | 1 | 0.0 | 2 | 1 | normal | Yes |
| 230 | 66 | 1 | asymptomatic | 112 | 212 | 0 | 2 | 132 | 1 | 0.1 | 1 | 1 | normal | Yes |
| 231 | 52 | 0 | nonanginal | 136 | 196 | 0 | 2 | 169 | 0 | 0.1 | 2 | 0 | normal | No |
| 232 | 55 | 0 | asymptomatic | 180 | 327 | 0 | 1 | 117 | 1 | 3.4 | 2 | 0 | normal | Yes |
| 233 | 49 | 1 | nonanginal | 118 | 149 | 0 | 2 | 126 | 0 | 0.8 | 1 | 3 | normal | Yes |
| 234 | 74 | 0 | nontypical | 120 | 269 | 0 | 2 | 121 | 1 | 0.2 | 1 | 1 | normal | No |
| 235 | 54 | 0 | nonanginal | 160 | 201 | 0 | 0 | 163 | 0 | 0.0 | 1 | 1 | normal | No |
| 236 | 54 | 1 | asymptomatic | 122 | 286 | 0 | 2 | 116 | 1 | 3.2 | 2 | 2 | normal | Yes |
| 237 | 56 | 1 | asymptomatic | 130 | 283 | 1 | 2 | 103 | 1 | 1.6 | 3 | 0 | reversable | Yes |
| 238 | 46 | 1 | asymptomatic | 120 | 249 | 0 | 2 | 144 | 0 | 0.8 | 1 | 0 | reversable | Yes |
| 239 | 49 | 0 | nontypical | 134 | 271 | 0 | 0 | 162 | 0 | 0.0 | 2 | 0 | normal | No |
| 240 | 42 | 1 | nontypical | 120 | 295 | 0 | 0 | 162 | 0 | 0.0 | 1 | 0 | normal | No |
| 241 | 41 | 1 | nontypical | 110 | 235 | 0 | 0 | 153 | 0 | 0.0 | 1 | 0 | normal | No |
| 242 | 41 | 0 | nontypical | 126 | 306 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 243 | 49 | 0 | asymptomatic | 130 | 269 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 244 | 61 | 1 | typical | 134 | 234 | 0 | 0 | 145 | 0 | 2.6 | 2 | 2 | normal | Yes |
| 245 | 60 | 0 | nonanginal | 120 | 178 | 1 | 0 | 96 | 0 | 0.0 | 1 | 0 | normal | No |
| 246 | 67 | 1 | asymptomatic | 120 | 237 | 0 | 0 | 71 | 0 | 1.0 | 2 | 0 | normal | Yes |
| 247 | 58 | 1 | asymptomatic | 100 | 234 | 0 | 0 | 156 | 0 | 0.1 | 1 | 1 | reversable | Yes |
| 248 | 47 | 1 | asymptomatic | 110 | 275 | 0 | 2 | 118 | 1 | 1.0 | 2 | 1 | normal | Yes |
| 249 | 52 | 1 | asymptomatic | 125 | 212 | 0 | 0 | 168 | 0 | 1.0 | 1 | 2 | reversable | Yes |
| 250 | 62 | 1 | nontypical | 128 | 208 | 1 | 2 | 140 | 0 | 0.0 | 1 | 0 | normal | No |
| 251 | 57 | 1 | asymptomatic | 110 | 201 | 0 | 0 | 126 | 1 | 1.5 | 2 | 0 | fixed | No |
| 252 | 58 | 1 | asymptomatic | 146 | 218 | 0 | 0 | 105 | 0 | 2.0 | 2 | 1 | reversable | Yes |
| 253 | 64 | 1 | asymptomatic | 128 | 263 | 0 | 0 | 105 | 1 | 0.2 | 2 | 1 | reversable | No |
| 254 | 51 | 0 | nonanginal | 120 | 295 | 0 | 2 | 157 | 0 | 0.6 | 1 | 0 | normal | No |
| 255 | 43 | 1 | asymptomatic | 115 | 303 | 0 | 0 | 181 | 0 | 1.2 | 2 | 0 | normal | No |
| 256 | 42 | 0 | nonanginal | 120 | 209 | 0 | 0 | 173 | 0 | 0.0 | 2 | 0 | normal | No |
| 257 | 67 | 0 | asymptomatic | 106 | 223 | 0 | 0 | 142 | 0 | 0.3 | 1 | 2 | normal | No |
| 258 | 76 | 0 | nonanginal | 140 | 197 | 0 | 1 | 116 | 0 | 1.1 | 2 | 0 | normal | No |
| 259 | 70 | 1 | nontypical | 156 | 245 | 0 | 2 | 143 | 0 | 0.0 | 1 | 0 | normal | No |
| 260 | 57 | 1 | nontypical | 124 | 261 | 0 | 0 | 141 | 0 | 0.3 | 1 | 0 | reversable | Yes |
| 261 | 44 | 0 | nonanginal | 118 | 242 | 0 | 0 | 149 | 0 | 0.3 | 2 | 1 | normal | No |
| 262 | 58 | 0 | nontypical | 136 | 319 | 1 | 2 | 152 | 0 | 0.0 | 1 | 2 | normal | Yes |
| 263 | 60 | 0 | typical | 150 | 240 | 0 | 0 | 171 | 0 | 0.9 | 1 | 0 | normal | No |
| 264 | 44 | 1 | nonanginal | 120 | 226 | 0 | 0 | 169 | 0 | 0.0 | 1 | 0 | normal | No |
| 265 | 61 | 1 | asymptomatic | 138 | 166 | 0 | 2 | 125 | 1 | 3.6 | 2 | 1 | normal | Yes |
| 266 | 42 | 1 | asymptomatic | 136 | 315 | 0 | 0 | 125 | 1 | 1.8 | 2 | 0 | fixed | Yes |
| 268 | 59 | 1 | nonanginal | 126 | 218 | 1 | 0 | 134 | 0 | 2.2 | 2 | 1 | fixed | Yes |
| 269 | 40 | 1 | asymptomatic | 152 | 223 | 0 | 0 | 181 | 0 | 0.0 | 1 | 0 | reversable | Yes |
| 270 | 42 | 1 | nonanginal | 130 | 180 | 0 | 0 | 150 | 0 | 0.0 | 1 | 0 | normal | No |
| 271 | 61 | 1 | asymptomatic | 140 | 207 | 0 | 2 | 138 | 1 | 1.9 | 1 | 1 | reversable | Yes |
| 272 | 66 | 1 | asymptomatic | 160 | 228 | 0 | 2 | 138 | 0 | 2.3 | 1 | 0 | fixed | No |
| 273 | 46 | 1 | asymptomatic | 140 | 311 | 0 | 0 | 120 | 1 | 1.8 | 2 | 2 | reversable | Yes |
| 274 | 71 | 0 | asymptomatic | 112 | 149 | 0 | 0 | 125 | 0 | 1.6 | 2 | 0 | normal | No |
| 275 | 59 | 1 | typical | 134 | 204 | 0 | 0 | 162 | 0 | 0.8 | 1 | 2 | normal | Yes |
| 276 | 64 | 1 | typical | 170 | 227 | 0 | 2 | 155 | 0 | 0.6 | 2 | 0 | reversable | No |
| 277 | 66 | 0 | nonanginal | 146 | 278 | 0 | 2 | 152 | 0 | 0.0 | 2 | 1 | normal | No |
| 278 | 39 | 0 | nonanginal | 138 | 220 | 0 | 0 | 152 | 0 | 0.0 | 2 | 0 | normal | No |
| 279 | 57 | 1 | nontypical | 154 | 232 | 0 | 2 | 164 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 280 | 58 | 0 | asymptomatic | 130 | 197 | 0 | 0 | 131 | 0 | 0.6 | 2 | 0 | normal | No |
| 281 | 57 | 1 | asymptomatic | 110 | 335 | 0 | 0 | 143 | 1 | 3.0 | 2 | 1 | reversable | Yes |
| 282 | 47 | 1 | nonanginal | 130 | 253 | 0 | 0 | 179 | 0 | 0.0 | 1 | 0 | normal | No |
| 283 | 55 | 0 | asymptomatic | 128 | 205 | 0 | 1 | 130 | 1 | 2.0 | 2 | 1 | reversable | Yes |
| 284 | 35 | 1 | nontypical | 122 | 192 | 0 | 0 | 174 | 0 | 0.0 | 1 | 0 | normal | No |
| 285 | 61 | 1 | asymptomatic | 148 | 203 | 0 | 0 | 161 | 0 | 0.0 | 1 | 1 | reversable | Yes |
| 286 | 58 | 1 | asymptomatic | 114 | 318 | 0 | 1 | 140 | 0 | 4.4 | 3 | 3 | fixed | Yes |
| 287 | 58 | 0 | asymptomatic | 170 | 225 | 1 | 2 | 146 | 1 | 2.8 | 2 | 2 | fixed | Yes |
| 289 | 56 | 1 | nontypical | 130 | 221 | 0 | 2 | 163 | 0 | 0.0 | 1 | 0 | reversable | No |
| 290 | 56 | 1 | nontypical | 120 | 240 | 0 | 0 | 169 | 0 | 0.0 | 3 | 0 | normal | No |
| 291 | 67 | 1 | nonanginal | 152 | 212 | 0 | 2 | 150 | 0 | 0.8 | 2 | 0 | reversable | Yes |
| 292 | 55 | 0 | nontypical | 132 | 342 | 0 | 0 | 166 | 0 | 1.2 | 1 | 0 | normal | No |
| 293 | 44 | 1 | asymptomatic | 120 | 169 | 0 | 0 | 144 | 1 | 2.8 | 3 | 0 | fixed | Yes |
| 294 | 63 | 1 | asymptomatic | 140 | 187 | 0 | 2 | 144 | 1 | 4.0 | 1 | 2 | reversable | Yes |
| 295 | 63 | 0 | asymptomatic | 124 | 197 | 0 | 0 | 136 | 1 | 0.0 | 2 | 0 | normal | Yes |
| 296 | 41 | 1 | nontypical | 120 | 157 | 0 | 0 | 182 | 0 | 0.0 | 1 | 0 | normal | No |
| 297 | 59 | 1 | asymptomatic | 164 | 176 | 1 | 2 | 90 | 0 | 1.0 | 2 | 2 | fixed | Yes |
| 298 | 57 | 0 | asymptomatic | 140 | 241 | 0 | 0 | 123 | 1 | 0.2 | 2 | 0 | reversable | Yes |
| 299 | 45 | 1 | typical | 110 | 264 | 0 | 0 | 132 | 0 | 1.2 | 2 | 0 | reversable | Yes |
| 300 | 68 | 1 | asymptomatic | 144 | 193 | 1 | 0 | 141 | 0 | 3.4 | 2 | 2 | reversable | Yes |
| 301 | 57 | 1 | asymptomatic | 130 | 131 | 0 | 0 | 115 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 302 | 57 | 0 | nontypical | 130 | 236 | 0 | 2 | 174 | 0 | 0.0 | 2 | 1 | normal | Yes |
16.7.2 CarSeats数据
| Sales | CompPrice | Income | Advertising | Population | Price | ShelveLoc | Age | Education | Urban | US |
|---|---|---|---|---|---|---|---|---|---|---|
| 9.50 | 138 | 73 | 11 | 276 | 120 | Bad | 42 | 17 | Yes | Yes |
| 11.22 | 111 | 48 | 16 | 260 | 83 | Good | 65 | 10 | Yes | Yes |
| 10.06 | 113 | 35 | 10 | 269 | 80 | Medium | 59 | 12 | Yes | Yes |
| 7.40 | 117 | 100 | 4 | 466 | 97 | Medium | 55 | 14 | Yes | Yes |
| 4.15 | 141 | 64 | 3 | 340 | 128 | Bad | 38 | 13 | Yes | No |
| 10.81 | 124 | 113 | 13 | 501 | 72 | Bad | 78 | 16 | No | Yes |
| 6.63 | 115 | 105 | 0 | 45 | 108 | Medium | 71 | 15 | Yes | No |
| 11.85 | 136 | 81 | 15 | 425 | 120 | Good | 67 | 10 | Yes | Yes |
| 6.54 | 132 | 110 | 0 | 108 | 124 | Medium | 76 | 10 | No | No |
| 4.69 | 132 | 113 | 0 | 131 | 124 | Medium | 76 | 17 | No | Yes |
| 9.01 | 121 | 78 | 9 | 150 | 100 | Bad | 26 | 10 | No | Yes |
| 11.96 | 117 | 94 | 4 | 503 | 94 | Good | 50 | 13 | Yes | Yes |
| 3.98 | 122 | 35 | 2 | 393 | 136 | Medium | 62 | 18 | Yes | No |
| 10.96 | 115 | 28 | 11 | 29 | 86 | Good | 53 | 18 | Yes | Yes |
| 11.17 | 107 | 117 | 11 | 148 | 118 | Good | 52 | 18 | Yes | Yes |
| 8.71 | 149 | 95 | 5 | 400 | 144 | Medium | 76 | 18 | No | No |
| 7.58 | 118 | 32 | 0 | 284 | 110 | Good | 63 | 13 | Yes | No |
| 12.29 | 147 | 74 | 13 | 251 | 131 | Good | 52 | 10 | Yes | Yes |
| 13.91 | 110 | 110 | 0 | 408 | 68 | Good | 46 | 17 | No | Yes |
| 8.73 | 129 | 76 | 16 | 58 | 121 | Medium | 69 | 12 | Yes | Yes |
| 6.41 | 125 | 90 | 2 | 367 | 131 | Medium | 35 | 18 | Yes | Yes |
| 12.13 | 134 | 29 | 12 | 239 | 109 | Good | 62 | 18 | No | Yes |
| 5.08 | 128 | 46 | 6 | 497 | 138 | Medium | 42 | 13 | Yes | No |
| 5.87 | 121 | 31 | 0 | 292 | 109 | Medium | 79 | 10 | Yes | No |
| 10.14 | 145 | 119 | 16 | 294 | 113 | Bad | 42 | 12 | Yes | Yes |
| 14.90 | 139 | 32 | 0 | 176 | 82 | Good | 54 | 11 | No | No |
| 8.33 | 107 | 115 | 11 | 496 | 131 | Good | 50 | 11 | No | Yes |
| 5.27 | 98 | 118 | 0 | 19 | 107 | Medium | 64 | 17 | Yes | No |
| 2.99 | 103 | 74 | 0 | 359 | 97 | Bad | 55 | 11 | Yes | Yes |
| 7.81 | 104 | 99 | 15 | 226 | 102 | Bad | 58 | 17 | Yes | Yes |
| 13.55 | 125 | 94 | 0 | 447 | 89 | Good | 30 | 12 | Yes | No |
| 8.25 | 136 | 58 | 16 | 241 | 131 | Medium | 44 | 18 | Yes | Yes |
| 6.20 | 107 | 32 | 12 | 236 | 137 | Good | 64 | 10 | No | Yes |
| 8.77 | 114 | 38 | 13 | 317 | 128 | Good | 50 | 16 | Yes | Yes |
| 2.67 | 115 | 54 | 0 | 406 | 128 | Medium | 42 | 17 | Yes | Yes |
| 11.07 | 131 | 84 | 11 | 29 | 96 | Medium | 44 | 17 | No | Yes |
| 8.89 | 122 | 76 | 0 | 270 | 100 | Good | 60 | 18 | No | No |
| 4.95 | 121 | 41 | 5 | 412 | 110 | Medium | 54 | 10 | Yes | Yes |
| 6.59 | 109 | 73 | 0 | 454 | 102 | Medium | 65 | 15 | Yes | No |
| 3.24 | 130 | 60 | 0 | 144 | 138 | Bad | 38 | 10 | No | No |
| 2.07 | 119 | 98 | 0 | 18 | 126 | Bad | 73 | 17 | No | No |
| 7.96 | 157 | 53 | 0 | 403 | 124 | Bad | 58 | 16 | Yes | No |
| 10.43 | 77 | 69 | 0 | 25 | 24 | Medium | 50 | 18 | Yes | No |
| 4.12 | 123 | 42 | 11 | 16 | 134 | Medium | 59 | 13 | Yes | Yes |
| 4.16 | 85 | 79 | 6 | 325 | 95 | Medium | 69 | 13 | Yes | Yes |
| 4.56 | 141 | 63 | 0 | 168 | 135 | Bad | 44 | 12 | Yes | Yes |
| 12.44 | 127 | 90 | 14 | 16 | 70 | Medium | 48 | 15 | No | Yes |
| 4.38 | 126 | 98 | 0 | 173 | 108 | Bad | 55 | 16 | Yes | No |
| 3.91 | 116 | 52 | 0 | 349 | 98 | Bad | 69 | 18 | Yes | No |
| 10.61 | 157 | 93 | 0 | 51 | 149 | Good | 32 | 17 | Yes | No |
| 1.42 | 99 | 32 | 18 | 341 | 108 | Bad | 80 | 16 | Yes | Yes |
| 4.42 | 121 | 90 | 0 | 150 | 108 | Bad | 75 | 16 | Yes | No |
| 7.91 | 153 | 40 | 3 | 112 | 129 | Bad | 39 | 18 | Yes | Yes |
| 6.92 | 109 | 64 | 13 | 39 | 119 | Medium | 61 | 17 | Yes | Yes |
| 4.90 | 134 | 103 | 13 | 25 | 144 | Medium | 76 | 17 | No | Yes |
| 6.85 | 143 | 81 | 5 | 60 | 154 | Medium | 61 | 18 | Yes | Yes |
| 11.91 | 133 | 82 | 0 | 54 | 84 | Medium | 50 | 17 | Yes | No |
| 0.91 | 93 | 91 | 0 | 22 | 117 | Bad | 75 | 11 | Yes | No |
| 5.42 | 103 | 93 | 15 | 188 | 103 | Bad | 74 | 16 | Yes | Yes |
| 5.21 | 118 | 71 | 4 | 148 | 114 | Medium | 80 | 13 | Yes | No |
| 8.32 | 122 | 102 | 19 | 469 | 123 | Bad | 29 | 13 | Yes | Yes |
| 7.32 | 105 | 32 | 0 | 358 | 107 | Medium | 26 | 13 | No | No |
| 1.82 | 139 | 45 | 0 | 146 | 133 | Bad | 77 | 17 | Yes | Yes |
| 8.47 | 119 | 88 | 10 | 170 | 101 | Medium | 61 | 13 | Yes | Yes |
| 7.80 | 100 | 67 | 12 | 184 | 104 | Medium | 32 | 16 | No | Yes |
| 4.90 | 122 | 26 | 0 | 197 | 128 | Medium | 55 | 13 | No | No |
| 8.85 | 127 | 92 | 0 | 508 | 91 | Medium | 56 | 18 | Yes | No |
| 9.01 | 126 | 61 | 14 | 152 | 115 | Medium | 47 | 16 | Yes | Yes |
| 13.39 | 149 | 69 | 20 | 366 | 134 | Good | 60 | 13 | Yes | Yes |
| 7.99 | 127 | 59 | 0 | 339 | 99 | Medium | 65 | 12 | Yes | No |
| 9.46 | 89 | 81 | 15 | 237 | 99 | Good | 74 | 12 | Yes | Yes |
| 6.50 | 148 | 51 | 16 | 148 | 150 | Medium | 58 | 17 | No | Yes |
| 5.52 | 115 | 45 | 0 | 432 | 116 | Medium | 25 | 15 | Yes | No |
| 12.61 | 118 | 90 | 10 | 54 | 104 | Good | 31 | 11 | No | Yes |
| 6.20 | 150 | 68 | 5 | 125 | 136 | Medium | 64 | 13 | No | Yes |
| 8.55 | 88 | 111 | 23 | 480 | 92 | Bad | 36 | 16 | No | Yes |
| 10.64 | 102 | 87 | 10 | 346 | 70 | Medium | 64 | 15 | Yes | Yes |
| 7.70 | 118 | 71 | 12 | 44 | 89 | Medium | 67 | 18 | No | Yes |
| 4.43 | 134 | 48 | 1 | 139 | 145 | Medium | 65 | 12 | Yes | Yes |
| 9.14 | 134 | 67 | 0 | 286 | 90 | Bad | 41 | 13 | Yes | No |
| 8.01 | 113 | 100 | 16 | 353 | 79 | Bad | 68 | 11 | Yes | Yes |
| 7.52 | 116 | 72 | 0 | 237 | 128 | Good | 70 | 13 | Yes | No |
| 11.62 | 151 | 83 | 4 | 325 | 139 | Good | 28 | 17 | Yes | Yes |
| 4.42 | 109 | 36 | 7 | 468 | 94 | Bad | 56 | 11 | Yes | Yes |
| 2.23 | 111 | 25 | 0 | 52 | 121 | Bad | 43 | 18 | No | No |
| 8.47 | 125 | 103 | 0 | 304 | 112 | Medium | 49 | 13 | No | No |
| 8.70 | 150 | 84 | 9 | 432 | 134 | Medium | 64 | 15 | Yes | No |
| 11.70 | 131 | 67 | 7 | 272 | 126 | Good | 54 | 16 | No | Yes |
| 6.56 | 117 | 42 | 7 | 144 | 111 | Medium | 62 | 10 | Yes | Yes |
| 7.95 | 128 | 66 | 3 | 493 | 119 | Medium | 45 | 16 | No | No |
| 5.33 | 115 | 22 | 0 | 491 | 103 | Medium | 64 | 11 | No | No |
| 4.81 | 97 | 46 | 11 | 267 | 107 | Medium | 80 | 15 | Yes | Yes |
| 4.53 | 114 | 113 | 0 | 97 | 125 | Medium | 29 | 12 | Yes | No |
| 8.86 | 145 | 30 | 0 | 67 | 104 | Medium | 55 | 17 | Yes | No |
| 8.39 | 115 | 97 | 5 | 134 | 84 | Bad | 55 | 11 | Yes | Yes |
| 5.58 | 134 | 25 | 10 | 237 | 148 | Medium | 59 | 13 | Yes | Yes |
| 9.48 | 147 | 42 | 10 | 407 | 132 | Good | 73 | 16 | No | Yes |
| 7.45 | 161 | 82 | 5 | 287 | 129 | Bad | 33 | 16 | Yes | Yes |
| 12.49 | 122 | 77 | 24 | 382 | 127 | Good | 36 | 16 | No | Yes |
| 4.88 | 121 | 47 | 3 | 220 | 107 | Bad | 56 | 16 | No | Yes |
| 4.11 | 113 | 69 | 11 | 94 | 106 | Medium | 76 | 12 | No | Yes |
| 6.20 | 128 | 93 | 0 | 89 | 118 | Medium | 34 | 18 | Yes | No |
| 5.30 | 113 | 22 | 0 | 57 | 97 | Medium | 65 | 16 | No | No |
| 5.07 | 123 | 91 | 0 | 334 | 96 | Bad | 78 | 17 | Yes | Yes |
| 4.62 | 121 | 96 | 0 | 472 | 138 | Medium | 51 | 12 | Yes | No |
| 5.55 | 104 | 100 | 8 | 398 | 97 | Medium | 61 | 11 | Yes | Yes |
| 0.16 | 102 | 33 | 0 | 217 | 139 | Medium | 70 | 18 | No | No |
| 8.55 | 134 | 107 | 0 | 104 | 108 | Medium | 60 | 12 | Yes | No |
| 3.47 | 107 | 79 | 2 | 488 | 103 | Bad | 65 | 16 | Yes | No |
| 8.98 | 115 | 65 | 0 | 217 | 90 | Medium | 60 | 17 | No | No |
| 9.00 | 128 | 62 | 7 | 125 | 116 | Medium | 43 | 14 | Yes | Yes |
| 6.62 | 132 | 118 | 12 | 272 | 151 | Medium | 43 | 14 | Yes | Yes |
| 6.67 | 116 | 99 | 5 | 298 | 125 | Good | 62 | 12 | Yes | Yes |
| 6.01 | 131 | 29 | 11 | 335 | 127 | Bad | 33 | 12 | Yes | Yes |
| 9.31 | 122 | 87 | 9 | 17 | 106 | Medium | 65 | 13 | Yes | Yes |
| 8.54 | 139 | 35 | 0 | 95 | 129 | Medium | 42 | 13 | Yes | No |
| 5.08 | 135 | 75 | 0 | 202 | 128 | Medium | 80 | 10 | No | No |
| 8.80 | 145 | 53 | 0 | 507 | 119 | Medium | 41 | 12 | Yes | No |
| 7.57 | 112 | 88 | 2 | 243 | 99 | Medium | 62 | 11 | Yes | Yes |
| 7.37 | 130 | 94 | 8 | 137 | 128 | Medium | 64 | 12 | Yes | Yes |
| 6.87 | 128 | 105 | 11 | 249 | 131 | Medium | 63 | 13 | Yes | Yes |
| 11.67 | 125 | 89 | 10 | 380 | 87 | Bad | 28 | 10 | Yes | Yes |
| 6.88 | 119 | 100 | 5 | 45 | 108 | Medium | 75 | 10 | Yes | Yes |
| 8.19 | 127 | 103 | 0 | 125 | 155 | Good | 29 | 15 | No | Yes |
| 8.87 | 131 | 113 | 0 | 181 | 120 | Good | 63 | 14 | Yes | No |
| 9.34 | 89 | 78 | 0 | 181 | 49 | Medium | 43 | 15 | No | No |
| 11.27 | 153 | 68 | 2 | 60 | 133 | Good | 59 | 16 | Yes | Yes |
| 6.52 | 125 | 48 | 3 | 192 | 116 | Medium | 51 | 14 | Yes | Yes |
| 4.96 | 133 | 100 | 3 | 350 | 126 | Bad | 55 | 13 | Yes | Yes |
| 4.47 | 143 | 120 | 7 | 279 | 147 | Bad | 40 | 10 | No | Yes |
| 8.41 | 94 | 84 | 13 | 497 | 77 | Medium | 51 | 12 | Yes | Yes |
| 6.50 | 108 | 69 | 3 | 208 | 94 | Medium | 77 | 16 | Yes | No |
| 9.54 | 125 | 87 | 9 | 232 | 136 | Good | 72 | 10 | Yes | Yes |
| 7.62 | 132 | 98 | 2 | 265 | 97 | Bad | 62 | 12 | Yes | Yes |
| 3.67 | 132 | 31 | 0 | 327 | 131 | Medium | 76 | 16 | Yes | No |
| 6.44 | 96 | 94 | 14 | 384 | 120 | Medium | 36 | 18 | No | Yes |
| 5.17 | 131 | 75 | 0 | 10 | 120 | Bad | 31 | 18 | No | No |
| 6.52 | 128 | 42 | 0 | 436 | 118 | Medium | 80 | 11 | Yes | No |
| 10.27 | 125 | 103 | 12 | 371 | 109 | Medium | 44 | 10 | Yes | Yes |
| 12.30 | 146 | 62 | 10 | 310 | 94 | Medium | 30 | 13 | No | Yes |
| 6.03 | 133 | 60 | 10 | 277 | 129 | Medium | 45 | 18 | Yes | Yes |
| 6.53 | 140 | 42 | 0 | 331 | 131 | Bad | 28 | 15 | Yes | No |
| 7.44 | 124 | 84 | 0 | 300 | 104 | Medium | 77 | 15 | Yes | No |
| 0.53 | 122 | 88 | 7 | 36 | 159 | Bad | 28 | 17 | Yes | Yes |
| 9.09 | 132 | 68 | 0 | 264 | 123 | Good | 34 | 11 | No | No |
| 8.77 | 144 | 63 | 11 | 27 | 117 | Medium | 47 | 17 | Yes | Yes |
| 3.90 | 114 | 83 | 0 | 412 | 131 | Bad | 39 | 14 | Yes | No |
| 10.51 | 140 | 54 | 9 | 402 | 119 | Good | 41 | 16 | No | Yes |
| 7.56 | 110 | 119 | 0 | 384 | 97 | Medium | 72 | 14 | No | Yes |
| 11.48 | 121 | 120 | 13 | 140 | 87 | Medium | 56 | 11 | Yes | Yes |
| 10.49 | 122 | 84 | 8 | 176 | 114 | Good | 57 | 10 | No | Yes |
| 10.77 | 111 | 58 | 17 | 407 | 103 | Good | 75 | 17 | No | Yes |
| 7.64 | 128 | 78 | 0 | 341 | 128 | Good | 45 | 13 | No | No |
| 5.93 | 150 | 36 | 7 | 488 | 150 | Medium | 25 | 17 | No | Yes |
| 6.89 | 129 | 69 | 10 | 289 | 110 | Medium | 50 | 16 | No | Yes |
| 7.71 | 98 | 72 | 0 | 59 | 69 | Medium | 65 | 16 | Yes | No |
| 7.49 | 146 | 34 | 0 | 220 | 157 | Good | 51 | 16 | Yes | No |
| 10.21 | 121 | 58 | 8 | 249 | 90 | Medium | 48 | 13 | No | Yes |
| 12.53 | 142 | 90 | 1 | 189 | 112 | Good | 39 | 10 | No | Yes |
| 9.32 | 119 | 60 | 0 | 372 | 70 | Bad | 30 | 18 | No | No |
| 4.67 | 111 | 28 | 0 | 486 | 111 | Medium | 29 | 12 | No | No |
| 2.93 | 143 | 21 | 5 | 81 | 160 | Medium | 67 | 12 | No | Yes |
| 3.63 | 122 | 74 | 0 | 424 | 149 | Medium | 51 | 13 | Yes | No |
| 5.68 | 130 | 64 | 0 | 40 | 106 | Bad | 39 | 17 | No | No |
| 8.22 | 148 | 64 | 0 | 58 | 141 | Medium | 27 | 13 | No | Yes |
| 0.37 | 147 | 58 | 7 | 100 | 191 | Bad | 27 | 15 | Yes | Yes |
| 6.71 | 119 | 67 | 17 | 151 | 137 | Medium | 55 | 11 | Yes | Yes |
| 6.71 | 106 | 73 | 0 | 216 | 93 | Medium | 60 | 13 | Yes | No |
| 7.30 | 129 | 89 | 0 | 425 | 117 | Medium | 45 | 10 | Yes | No |
| 11.48 | 104 | 41 | 15 | 492 | 77 | Good | 73 | 18 | Yes | Yes |
| 8.01 | 128 | 39 | 12 | 356 | 118 | Medium | 71 | 10 | Yes | Yes |
| 12.49 | 93 | 106 | 12 | 416 | 55 | Medium | 75 | 15 | Yes | Yes |
| 9.03 | 104 | 102 | 13 | 123 | 110 | Good | 35 | 16 | Yes | Yes |
| 6.38 | 135 | 91 | 5 | 207 | 128 | Medium | 66 | 18 | Yes | Yes |
| 0.00 | 139 | 24 | 0 | 358 | 185 | Medium | 79 | 15 | No | No |
| 7.54 | 115 | 89 | 0 | 38 | 122 | Medium | 25 | 12 | Yes | No |
| 5.61 | 138 | 107 | 9 | 480 | 154 | Medium | 47 | 11 | No | Yes |
| 10.48 | 138 | 72 | 0 | 148 | 94 | Medium | 27 | 17 | Yes | Yes |
| 10.66 | 104 | 71 | 14 | 89 | 81 | Medium | 25 | 14 | No | Yes |
| 7.78 | 144 | 25 | 3 | 70 | 116 | Medium | 77 | 18 | Yes | Yes |
| 4.94 | 137 | 112 | 15 | 434 | 149 | Bad | 66 | 13 | Yes | Yes |
| 7.43 | 121 | 83 | 0 | 79 | 91 | Medium | 68 | 11 | Yes | No |
| 4.74 | 137 | 60 | 4 | 230 | 140 | Bad | 25 | 13 | Yes | No |
| 5.32 | 118 | 74 | 6 | 426 | 102 | Medium | 80 | 18 | Yes | Yes |
| 9.95 | 132 | 33 | 7 | 35 | 97 | Medium | 60 | 11 | No | Yes |
| 10.07 | 130 | 100 | 11 | 449 | 107 | Medium | 64 | 10 | Yes | Yes |
| 8.68 | 120 | 51 | 0 | 93 | 86 | Medium | 46 | 17 | No | No |
| 6.03 | 117 | 32 | 0 | 142 | 96 | Bad | 62 | 17 | Yes | No |
| 8.07 | 116 | 37 | 0 | 426 | 90 | Medium | 76 | 15 | Yes | No |
| 12.11 | 118 | 117 | 18 | 509 | 104 | Medium | 26 | 15 | No | Yes |
| 8.79 | 130 | 37 | 13 | 297 | 101 | Medium | 37 | 13 | No | Yes |
| 6.67 | 156 | 42 | 13 | 170 | 173 | Good | 74 | 14 | Yes | Yes |
| 7.56 | 108 | 26 | 0 | 408 | 93 | Medium | 56 | 14 | No | No |
| 13.28 | 139 | 70 | 7 | 71 | 96 | Good | 61 | 10 | Yes | Yes |
| 7.23 | 112 | 98 | 18 | 481 | 128 | Medium | 45 | 11 | Yes | Yes |
| 4.19 | 117 | 93 | 4 | 420 | 112 | Bad | 66 | 11 | Yes | Yes |
| 4.10 | 130 | 28 | 6 | 410 | 133 | Bad | 72 | 16 | Yes | Yes |
| 2.52 | 124 | 61 | 0 | 333 | 138 | Medium | 76 | 16 | Yes | No |
| 3.62 | 112 | 80 | 5 | 500 | 128 | Medium | 69 | 10 | Yes | Yes |
| 6.42 | 122 | 88 | 5 | 335 | 126 | Medium | 64 | 14 | Yes | Yes |
| 5.56 | 144 | 92 | 0 | 349 | 146 | Medium | 62 | 12 | No | No |
| 5.94 | 138 | 83 | 0 | 139 | 134 | Medium | 54 | 18 | Yes | No |
| 4.10 | 121 | 78 | 4 | 413 | 130 | Bad | 46 | 10 | No | Yes |
| 2.05 | 131 | 82 | 0 | 132 | 157 | Bad | 25 | 14 | Yes | No |
| 8.74 | 155 | 80 | 0 | 237 | 124 | Medium | 37 | 14 | Yes | No |
| 5.68 | 113 | 22 | 1 | 317 | 132 | Medium | 28 | 12 | Yes | No |
| 4.97 | 162 | 67 | 0 | 27 | 160 | Medium | 77 | 17 | Yes | Yes |
| 8.19 | 111 | 105 | 0 | 466 | 97 | Bad | 61 | 10 | No | No |
| 7.78 | 86 | 54 | 0 | 497 | 64 | Bad | 33 | 12 | Yes | No |
| 3.02 | 98 | 21 | 11 | 326 | 90 | Bad | 76 | 11 | No | Yes |
| 4.36 | 125 | 41 | 2 | 357 | 123 | Bad | 47 | 14 | No | Yes |
| 9.39 | 117 | 118 | 14 | 445 | 120 | Medium | 32 | 15 | Yes | Yes |
| 12.04 | 145 | 69 | 19 | 501 | 105 | Medium | 45 | 11 | Yes | Yes |
| 8.23 | 149 | 84 | 5 | 220 | 139 | Medium | 33 | 10 | Yes | Yes |
| 4.83 | 115 | 115 | 3 | 48 | 107 | Medium | 73 | 18 | Yes | Yes |
| 2.34 | 116 | 83 | 15 | 170 | 144 | Bad | 71 | 11 | Yes | Yes |
| 5.73 | 141 | 33 | 0 | 243 | 144 | Medium | 34 | 17 | Yes | No |
| 4.34 | 106 | 44 | 0 | 481 | 111 | Medium | 70 | 14 | No | No |
| 9.70 | 138 | 61 | 12 | 156 | 120 | Medium | 25 | 14 | Yes | Yes |
| 10.62 | 116 | 79 | 19 | 359 | 116 | Good | 58 | 17 | Yes | Yes |
| 10.59 | 131 | 120 | 15 | 262 | 124 | Medium | 30 | 10 | Yes | Yes |
| 6.43 | 124 | 44 | 0 | 125 | 107 | Medium | 80 | 11 | Yes | No |
| 7.49 | 136 | 119 | 6 | 178 | 145 | Medium | 35 | 13 | Yes | Yes |
| 3.45 | 110 | 45 | 9 | 276 | 125 | Medium | 62 | 14 | Yes | Yes |
| 4.10 | 134 | 82 | 0 | 464 | 141 | Medium | 48 | 13 | No | No |
| 6.68 | 107 | 25 | 0 | 412 | 82 | Bad | 36 | 14 | Yes | No |
| 7.80 | 119 | 33 | 0 | 245 | 122 | Good | 56 | 14 | Yes | No |
| 8.69 | 113 | 64 | 10 | 68 | 101 | Medium | 57 | 16 | Yes | Yes |
| 5.40 | 149 | 73 | 13 | 381 | 163 | Bad | 26 | 11 | No | Yes |
| 11.19 | 98 | 104 | 0 | 404 | 72 | Medium | 27 | 18 | No | No |
| 5.16 | 115 | 60 | 0 | 119 | 114 | Bad | 38 | 14 | No | No |
| 8.09 | 132 | 69 | 0 | 123 | 122 | Medium | 27 | 11 | No | No |
| 13.14 | 137 | 80 | 10 | 24 | 105 | Good | 61 | 15 | Yes | Yes |
| 8.65 | 123 | 76 | 18 | 218 | 120 | Medium | 29 | 14 | No | Yes |
| 9.43 | 115 | 62 | 11 | 289 | 129 | Good | 56 | 16 | No | Yes |
| 5.53 | 126 | 32 | 8 | 95 | 132 | Medium | 50 | 17 | Yes | Yes |
| 9.32 | 141 | 34 | 16 | 361 | 108 | Medium | 69 | 10 | Yes | Yes |
| 9.62 | 151 | 28 | 8 | 499 | 135 | Medium | 48 | 10 | Yes | Yes |
| 7.36 | 121 | 24 | 0 | 200 | 133 | Good | 73 | 13 | Yes | No |
| 3.89 | 123 | 105 | 0 | 149 | 118 | Bad | 62 | 16 | Yes | Yes |
| 10.31 | 159 | 80 | 0 | 362 | 121 | Medium | 26 | 18 | Yes | No |
| 12.01 | 136 | 63 | 0 | 160 | 94 | Medium | 38 | 12 | Yes | No |
| 4.68 | 124 | 46 | 0 | 199 | 135 | Medium | 52 | 14 | No | No |
| 7.82 | 124 | 25 | 13 | 87 | 110 | Medium | 57 | 10 | Yes | Yes |
| 8.78 | 130 | 30 | 0 | 391 | 100 | Medium | 26 | 18 | Yes | No |
| 10.00 | 114 | 43 | 0 | 199 | 88 | Good | 57 | 10 | No | Yes |
| 6.90 | 120 | 56 | 20 | 266 | 90 | Bad | 78 | 18 | Yes | Yes |
| 5.04 | 123 | 114 | 0 | 298 | 151 | Bad | 34 | 16 | Yes | No |
| 5.36 | 111 | 52 | 0 | 12 | 101 | Medium | 61 | 11 | Yes | Yes |
| 5.05 | 125 | 67 | 0 | 86 | 117 | Bad | 65 | 11 | Yes | No |
| 9.16 | 137 | 105 | 10 | 435 | 156 | Good | 72 | 14 | Yes | Yes |
| 3.72 | 139 | 111 | 5 | 310 | 132 | Bad | 62 | 13 | Yes | Yes |
| 8.31 | 133 | 97 | 0 | 70 | 117 | Medium | 32 | 16 | Yes | No |
| 5.64 | 124 | 24 | 5 | 288 | 122 | Medium | 57 | 12 | No | Yes |
| 9.58 | 108 | 104 | 23 | 353 | 129 | Good | 37 | 17 | Yes | Yes |
| 7.71 | 123 | 81 | 8 | 198 | 81 | Bad | 80 | 15 | Yes | Yes |
| 4.20 | 147 | 40 | 0 | 277 | 144 | Medium | 73 | 10 | Yes | No |
| 8.67 | 125 | 62 | 14 | 477 | 112 | Medium | 80 | 13 | Yes | Yes |
| 3.47 | 108 | 38 | 0 | 251 | 81 | Bad | 72 | 14 | No | No |
| 5.12 | 123 | 36 | 10 | 467 | 100 | Bad | 74 | 11 | No | Yes |
| 7.67 | 129 | 117 | 8 | 400 | 101 | Bad | 36 | 10 | Yes | Yes |
| 5.71 | 121 | 42 | 4 | 188 | 118 | Medium | 54 | 15 | Yes | Yes |
| 6.37 | 120 | 77 | 15 | 86 | 132 | Medium | 48 | 18 | Yes | Yes |
| 7.77 | 116 | 26 | 6 | 434 | 115 | Medium | 25 | 17 | Yes | Yes |
| 6.95 | 128 | 29 | 5 | 324 | 159 | Good | 31 | 15 | Yes | Yes |
| 5.31 | 130 | 35 | 10 | 402 | 129 | Bad | 39 | 17 | Yes | Yes |
| 9.10 | 128 | 93 | 12 | 343 | 112 | Good | 73 | 17 | No | Yes |
| 5.83 | 134 | 82 | 7 | 473 | 112 | Bad | 51 | 12 | No | Yes |
| 6.53 | 123 | 57 | 0 | 66 | 105 | Medium | 39 | 11 | Yes | No |
| 5.01 | 159 | 69 | 0 | 438 | 166 | Medium | 46 | 17 | Yes | No |
| 11.99 | 119 | 26 | 0 | 284 | 89 | Good | 26 | 10 | Yes | No |
| 4.55 | 111 | 56 | 0 | 504 | 110 | Medium | 62 | 16 | Yes | No |
| 12.98 | 113 | 33 | 0 | 14 | 63 | Good | 38 | 12 | Yes | No |
| 10.04 | 116 | 106 | 8 | 244 | 86 | Medium | 58 | 12 | Yes | Yes |
| 7.22 | 135 | 93 | 2 | 67 | 119 | Medium | 34 | 11 | Yes | Yes |
| 6.67 | 107 | 119 | 11 | 210 | 132 | Medium | 53 | 11 | Yes | Yes |
| 6.93 | 135 | 69 | 14 | 296 | 130 | Medium | 73 | 15 | Yes | Yes |
| 7.80 | 136 | 48 | 12 | 326 | 125 | Medium | 36 | 16 | Yes | Yes |
| 7.22 | 114 | 113 | 2 | 129 | 151 | Good | 40 | 15 | No | Yes |
| 3.42 | 141 | 57 | 13 | 376 | 158 | Medium | 64 | 18 | Yes | Yes |
| 2.86 | 121 | 86 | 10 | 496 | 145 | Bad | 51 | 10 | Yes | Yes |
| 11.19 | 122 | 69 | 7 | 303 | 105 | Good | 45 | 16 | No | Yes |
| 7.74 | 150 | 96 | 0 | 80 | 154 | Good | 61 | 11 | Yes | No |
| 5.36 | 135 | 110 | 0 | 112 | 117 | Medium | 80 | 16 | No | No |
| 6.97 | 106 | 46 | 11 | 414 | 96 | Bad | 79 | 17 | No | No |
| 7.60 | 146 | 26 | 11 | 261 | 131 | Medium | 39 | 10 | Yes | Yes |
| 7.53 | 117 | 118 | 11 | 429 | 113 | Medium | 67 | 18 | No | Yes |
| 6.88 | 95 | 44 | 4 | 208 | 72 | Bad | 44 | 17 | Yes | Yes |
| 6.98 | 116 | 40 | 0 | 74 | 97 | Medium | 76 | 15 | No | No |
| 8.75 | 143 | 77 | 25 | 448 | 156 | Medium | 43 | 17 | Yes | Yes |
| 9.49 | 107 | 111 | 14 | 400 | 103 | Medium | 41 | 11 | No | Yes |
| 6.64 | 118 | 70 | 0 | 106 | 89 | Bad | 39 | 17 | Yes | No |
| 11.82 | 113 | 66 | 16 | 322 | 74 | Good | 76 | 15 | Yes | Yes |
| 11.28 | 123 | 84 | 0 | 74 | 89 | Good | 59 | 10 | Yes | No |
| 12.66 | 148 | 76 | 3 | 126 | 99 | Good | 60 | 11 | Yes | Yes |
| 4.21 | 118 | 35 | 14 | 502 | 137 | Medium | 79 | 10 | No | Yes |
| 8.21 | 127 | 44 | 13 | 160 | 123 | Good | 63 | 18 | Yes | Yes |
| 3.07 | 118 | 83 | 13 | 276 | 104 | Bad | 75 | 10 | Yes | Yes |
| 10.98 | 148 | 63 | 0 | 312 | 130 | Good | 63 | 15 | Yes | No |
| 9.40 | 135 | 40 | 17 | 497 | 96 | Medium | 54 | 17 | No | Yes |
| 8.57 | 116 | 78 | 1 | 158 | 99 | Medium | 45 | 11 | Yes | Yes |
| 7.41 | 99 | 93 | 0 | 198 | 87 | Medium | 57 | 16 | Yes | Yes |
| 5.28 | 108 | 77 | 13 | 388 | 110 | Bad | 74 | 14 | Yes | Yes |
| 10.01 | 133 | 52 | 16 | 290 | 99 | Medium | 43 | 11 | Yes | Yes |
| 11.93 | 123 | 98 | 12 | 408 | 134 | Good | 29 | 10 | Yes | Yes |
| 8.03 | 115 | 29 | 26 | 394 | 132 | Medium | 33 | 13 | Yes | Yes |
| 4.78 | 131 | 32 | 1 | 85 | 133 | Medium | 48 | 12 | Yes | Yes |
| 5.90 | 138 | 92 | 0 | 13 | 120 | Bad | 61 | 12 | Yes | No |
| 9.24 | 126 | 80 | 19 | 436 | 126 | Medium | 52 | 10 | Yes | Yes |
| 11.18 | 131 | 111 | 13 | 33 | 80 | Bad | 68 | 18 | Yes | Yes |
| 9.53 | 175 | 65 | 29 | 419 | 166 | Medium | 53 | 12 | Yes | Yes |
| 6.15 | 146 | 68 | 12 | 328 | 132 | Bad | 51 | 14 | Yes | Yes |
| 6.80 | 137 | 117 | 5 | 337 | 135 | Bad | 38 | 10 | Yes | Yes |
| 9.33 | 103 | 81 | 3 | 491 | 54 | Medium | 66 | 13 | Yes | No |
| 7.72 | 133 | 33 | 10 | 333 | 129 | Good | 71 | 14 | Yes | Yes |
| 6.39 | 131 | 21 | 8 | 220 | 171 | Good | 29 | 14 | Yes | Yes |
| 15.63 | 122 | 36 | 5 | 369 | 72 | Good | 35 | 10 | Yes | Yes |
| 6.41 | 142 | 30 | 0 | 472 | 136 | Good | 80 | 15 | No | No |
| 10.08 | 116 | 72 | 10 | 456 | 130 | Good | 41 | 14 | No | Yes |
| 6.97 | 127 | 45 | 19 | 459 | 129 | Medium | 57 | 11 | No | Yes |
| 5.86 | 136 | 70 | 12 | 171 | 152 | Medium | 44 | 18 | Yes | Yes |
| 7.52 | 123 | 39 | 5 | 499 | 98 | Medium | 34 | 15 | Yes | No |
| 9.16 | 140 | 50 | 10 | 300 | 139 | Good | 60 | 15 | Yes | Yes |
| 10.36 | 107 | 105 | 18 | 428 | 103 | Medium | 34 | 12 | Yes | Yes |
| 2.66 | 136 | 65 | 4 | 133 | 150 | Bad | 53 | 13 | Yes | Yes |
| 11.70 | 144 | 69 | 11 | 131 | 104 | Medium | 47 | 11 | Yes | Yes |
| 4.69 | 133 | 30 | 0 | 152 | 122 | Medium | 53 | 17 | Yes | No |
| 6.23 | 112 | 38 | 17 | 316 | 104 | Medium | 80 | 16 | Yes | Yes |
| 3.15 | 117 | 66 | 1 | 65 | 111 | Bad | 55 | 11 | Yes | Yes |
| 11.27 | 100 | 54 | 9 | 433 | 89 | Good | 45 | 12 | Yes | Yes |
| 4.99 | 122 | 59 | 0 | 501 | 112 | Bad | 32 | 14 | No | No |
| 10.10 | 135 | 63 | 15 | 213 | 134 | Medium | 32 | 10 | Yes | Yes |
| 5.74 | 106 | 33 | 20 | 354 | 104 | Medium | 61 | 12 | Yes | Yes |
| 5.87 | 136 | 60 | 7 | 303 | 147 | Medium | 41 | 10 | Yes | Yes |
| 7.63 | 93 | 117 | 9 | 489 | 83 | Bad | 42 | 13 | Yes | Yes |
| 6.18 | 120 | 70 | 15 | 464 | 110 | Medium | 72 | 15 | Yes | Yes |
| 5.17 | 138 | 35 | 6 | 60 | 143 | Bad | 28 | 18 | Yes | No |
| 8.61 | 130 | 38 | 0 | 283 | 102 | Medium | 80 | 15 | Yes | No |
| 5.97 | 112 | 24 | 0 | 164 | 101 | Medium | 45 | 11 | Yes | No |
| 11.54 | 134 | 44 | 4 | 219 | 126 | Good | 44 | 15 | Yes | Yes |
| 7.50 | 140 | 29 | 0 | 105 | 91 | Bad | 43 | 16 | Yes | No |
| 7.38 | 98 | 120 | 0 | 268 | 93 | Medium | 72 | 10 | No | No |
| 7.81 | 137 | 102 | 13 | 422 | 118 | Medium | 71 | 10 | No | Yes |
| 5.99 | 117 | 42 | 10 | 371 | 121 | Bad | 26 | 14 | Yes | Yes |
| 8.43 | 138 | 80 | 0 | 108 | 126 | Good | 70 | 13 | No | Yes |
| 4.81 | 121 | 68 | 0 | 279 | 149 | Good | 79 | 12 | Yes | No |
| 8.97 | 132 | 107 | 0 | 144 | 125 | Medium | 33 | 13 | No | No |
| 6.88 | 96 | 39 | 0 | 161 | 112 | Good | 27 | 14 | No | No |
| 12.57 | 132 | 102 | 20 | 459 | 107 | Good | 49 | 11 | Yes | Yes |
| 9.32 | 134 | 27 | 18 | 467 | 96 | Medium | 49 | 14 | No | Yes |
| 8.64 | 111 | 101 | 17 | 266 | 91 | Medium | 63 | 17 | No | Yes |
| 10.44 | 124 | 115 | 16 | 458 | 105 | Medium | 62 | 16 | No | Yes |
| 13.44 | 133 | 103 | 14 | 288 | 122 | Good | 61 | 17 | Yes | Yes |
| 9.45 | 107 | 67 | 12 | 430 | 92 | Medium | 35 | 12 | No | Yes |
| 5.30 | 133 | 31 | 1 | 80 | 145 | Medium | 42 | 18 | Yes | Yes |
| 7.02 | 130 | 100 | 0 | 306 | 146 | Good | 42 | 11 | Yes | No |
| 3.58 | 142 | 109 | 0 | 111 | 164 | Good | 72 | 12 | Yes | No |
| 13.36 | 103 | 73 | 3 | 276 | 72 | Medium | 34 | 15 | Yes | Yes |
| 4.17 | 123 | 96 | 10 | 71 | 118 | Bad | 69 | 11 | Yes | Yes |
| 3.13 | 130 | 62 | 11 | 396 | 130 | Bad | 66 | 14 | Yes | Yes |
| 8.77 | 118 | 86 | 7 | 265 | 114 | Good | 52 | 15 | No | Yes |
| 8.68 | 131 | 25 | 10 | 183 | 104 | Medium | 56 | 15 | No | Yes |
| 5.25 | 131 | 55 | 0 | 26 | 110 | Bad | 79 | 12 | Yes | Yes |
| 10.26 | 111 | 75 | 1 | 377 | 108 | Good | 25 | 12 | Yes | No |
| 10.50 | 122 | 21 | 16 | 488 | 131 | Good | 30 | 14 | Yes | Yes |
| 6.53 | 154 | 30 | 0 | 122 | 162 | Medium | 57 | 17 | No | No |
| 5.98 | 124 | 56 | 11 | 447 | 134 | Medium | 53 | 12 | No | Yes |
| 14.37 | 95 | 106 | 0 | 256 | 53 | Good | 52 | 17 | Yes | No |
| 10.71 | 109 | 22 | 10 | 348 | 79 | Good | 74 | 14 | No | Yes |
| 10.26 | 135 | 100 | 22 | 463 | 122 | Medium | 36 | 14 | Yes | Yes |
| 7.68 | 126 | 41 | 22 | 403 | 119 | Bad | 42 | 12 | Yes | Yes |
| 9.08 | 152 | 81 | 0 | 191 | 126 | Medium | 54 | 16 | Yes | No |
| 7.80 | 121 | 50 | 0 | 508 | 98 | Medium | 65 | 11 | No | No |
| 5.58 | 137 | 71 | 0 | 402 | 116 | Medium | 78 | 17 | Yes | No |
| 9.44 | 131 | 47 | 7 | 90 | 118 | Medium | 47 | 12 | Yes | Yes |
| 7.90 | 132 | 46 | 4 | 206 | 124 | Medium | 73 | 11 | Yes | No |
| 16.27 | 141 | 60 | 19 | 319 | 92 | Good | 44 | 11 | Yes | Yes |
| 6.81 | 132 | 61 | 0 | 263 | 125 | Medium | 41 | 12 | No | No |
| 6.11 | 133 | 88 | 3 | 105 | 119 | Medium | 79 | 12 | Yes | Yes |
| 5.81 | 125 | 111 | 0 | 404 | 107 | Bad | 54 | 15 | Yes | No |
| 9.64 | 106 | 64 | 10 | 17 | 89 | Medium | 68 | 17 | Yes | Yes |
| 3.90 | 124 | 65 | 21 | 496 | 151 | Bad | 77 | 13 | Yes | Yes |
| 4.95 | 121 | 28 | 19 | 315 | 121 | Medium | 66 | 14 | Yes | Yes |
| 9.35 | 98 | 117 | 0 | 76 | 68 | Medium | 63 | 10 | Yes | No |
| 12.85 | 123 | 37 | 15 | 348 | 112 | Good | 28 | 12 | Yes | Yes |
| 5.87 | 131 | 73 | 13 | 455 | 132 | Medium | 62 | 17 | Yes | Yes |
| 5.32 | 152 | 116 | 0 | 170 | 160 | Medium | 39 | 16 | Yes | No |
| 8.67 | 142 | 73 | 14 | 238 | 115 | Medium | 73 | 14 | No | Yes |
| 8.14 | 135 | 89 | 11 | 245 | 78 | Bad | 79 | 16 | Yes | Yes |
| 8.44 | 128 | 42 | 8 | 328 | 107 | Medium | 35 | 12 | Yes | Yes |
| 5.47 | 108 | 75 | 9 | 61 | 111 | Medium | 67 | 12 | Yes | Yes |
| 6.10 | 153 | 63 | 0 | 49 | 124 | Bad | 56 | 16 | Yes | No |
| 4.53 | 129 | 42 | 13 | 315 | 130 | Bad | 34 | 13 | Yes | Yes |
| 5.57 | 109 | 51 | 10 | 26 | 120 | Medium | 30 | 17 | No | Yes |
| 5.35 | 130 | 58 | 19 | 366 | 139 | Bad | 33 | 16 | Yes | Yes |
| 12.57 | 138 | 108 | 17 | 203 | 128 | Good | 33 | 14 | Yes | Yes |
| 6.14 | 139 | 23 | 3 | 37 | 120 | Medium | 55 | 11 | No | Yes |
| 7.41 | 162 | 26 | 12 | 368 | 159 | Medium | 40 | 18 | Yes | Yes |
| 5.94 | 100 | 79 | 7 | 284 | 95 | Bad | 50 | 12 | Yes | Yes |
| 9.71 | 134 | 37 | 0 | 27 | 120 | Good | 49 | 16 | Yes | Yes |
16.7.3 Boston数据
| crim | zn | indus | chas | nox | rm | age | dis | rad | tax | ptratio | black | lstat | medv |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.00632 | 18.0 | 2.31 | 0 | 0.5380 | 6.575 | 65.2 | 4.0900 | 1 | 296 | 15.3 | 396.90 | 4.98 | 24.0 |
| 0.02731 | 0.0 | 7.07 | 0 | 0.4690 | 6.421 | 78.9 | 4.9671 | 2 | 242 | 17.8 | 396.90 | 9.14 | 21.6 |
| 0.02729 | 0.0 | 7.07 | 0 | 0.4690 | 7.185 | 61.1 | 4.9671 | 2 | 242 | 17.8 | 392.83 | 4.03 | 34.7 |
| 0.03237 | 0.0 | 2.18 | 0 | 0.4580 | 6.998 | 45.8 | 6.0622 | 3 | 222 | 18.7 | 394.63 | 2.94 | 33.4 |
| 0.06905 | 0.0 | 2.18 | 0 | 0.4580 | 7.147 | 54.2 | 6.0622 | 3 | 222 | 18.7 | 396.90 | 5.33 | 36.2 |
| 0.02985 | 0.0 | 2.18 | 0 | 0.4580 | 6.430 | 58.7 | 6.0622 | 3 | 222 | 18.7 | 394.12 | 5.21 | 28.7 |
| 0.08829 | 12.5 | 7.87 | 0 | 0.5240 | 6.012 | 66.6 | 5.5605 | 5 | 311 | 15.2 | 395.60 | 12.43 | 22.9 |
| 0.14455 | 12.5 | 7.87 | 0 | 0.5240 | 6.172 | 96.1 | 5.9505 | 5 | 311 | 15.2 | 396.90 | 19.15 | 27.1 |
| 0.21124 | 12.5 | 7.87 | 0 | 0.5240 | 5.631 | 100.0 | 6.0821 | 5 | 311 | 15.2 | 386.63 | 29.93 | 16.5 |
| 0.17004 | 12.5 | 7.87 | 0 | 0.5240 | 6.004 | 85.9 | 6.5921 | 5 | 311 | 15.2 | 386.71 | 17.10 | 18.9 |
| 0.22489 | 12.5 | 7.87 | 0 | 0.5240 | 6.377 | 94.3 | 6.3467 | 5 | 311 | 15.2 | 392.52 | 20.45 | 15.0 |
| 0.11747 | 12.5 | 7.87 | 0 | 0.5240 | 6.009 | 82.9 | 6.2267 | 5 | 311 | 15.2 | 396.90 | 13.27 | 18.9 |
| 0.09378 | 12.5 | 7.87 | 0 | 0.5240 | 5.889 | 39.0 | 5.4509 | 5 | 311 | 15.2 | 390.50 | 15.71 | 21.7 |
| 0.62976 | 0.0 | 8.14 | 0 | 0.5380 | 5.949 | 61.8 | 4.7075 | 4 | 307 | 21.0 | 396.90 | 8.26 | 20.4 |
| 0.63796 | 0.0 | 8.14 | 0 | 0.5380 | 6.096 | 84.5 | 4.4619 | 4 | 307 | 21.0 | 380.02 | 10.26 | 18.2 |
| 0.62739 | 0.0 | 8.14 | 0 | 0.5380 | 5.834 | 56.5 | 4.4986 | 4 | 307 | 21.0 | 395.62 | 8.47 | 19.9 |
| 1.05393 | 0.0 | 8.14 | 0 | 0.5380 | 5.935 | 29.3 | 4.4986 | 4 | 307 | 21.0 | 386.85 | 6.58 | 23.1 |
| 0.78420 | 0.0 | 8.14 | 0 | 0.5380 | 5.990 | 81.7 | 4.2579 | 4 | 307 | 21.0 | 386.75 | 14.67 | 17.5 |
| 0.80271 | 0.0 | 8.14 | 0 | 0.5380 | 5.456 | 36.6 | 3.7965 | 4 | 307 | 21.0 | 288.99 | 11.69 | 20.2 |
| 0.72580 | 0.0 | 8.14 | 0 | 0.5380 | 5.727 | 69.5 | 3.7965 | 4 | 307 | 21.0 | 390.95 | 11.28 | 18.2 |
| 1.25179 | 0.0 | 8.14 | 0 | 0.5380 | 5.570 | 98.1 | 3.7979 | 4 | 307 | 21.0 | 376.57 | 21.02 | 13.6 |
| 0.85204 | 0.0 | 8.14 | 0 | 0.5380 | 5.965 | 89.2 | 4.0123 | 4 | 307 | 21.0 | 392.53 | 13.83 | 19.6 |
| 1.23247 | 0.0 | 8.14 | 0 | 0.5380 | 6.142 | 91.7 | 3.9769 | 4 | 307 | 21.0 | 396.90 | 18.72 | 15.2 |
| 0.98843 | 0.0 | 8.14 | 0 | 0.5380 | 5.813 | 100.0 | 4.0952 | 4 | 307 | 21.0 | 394.54 | 19.88 | 14.5 |
| 0.75026 | 0.0 | 8.14 | 0 | 0.5380 | 5.924 | 94.1 | 4.3996 | 4 | 307 | 21.0 | 394.33 | 16.30 | 15.6 |
| 0.84054 | 0.0 | 8.14 | 0 | 0.5380 | 5.599 | 85.7 | 4.4546 | 4 | 307 | 21.0 | 303.42 | 16.51 | 13.9 |
| 0.67191 | 0.0 | 8.14 | 0 | 0.5380 | 5.813 | 90.3 | 4.6820 | 4 | 307 | 21.0 | 376.88 | 14.81 | 16.6 |
| 0.95577 | 0.0 | 8.14 | 0 | 0.5380 | 6.047 | 88.8 | 4.4534 | 4 | 307 | 21.0 | 306.38 | 17.28 | 14.8 |
| 0.77299 | 0.0 | 8.14 | 0 | 0.5380 | 6.495 | 94.4 | 4.4547 | 4 | 307 | 21.0 | 387.94 | 12.80 | 18.4 |
| 1.00245 | 0.0 | 8.14 | 0 | 0.5380 | 6.674 | 87.3 | 4.2390 | 4 | 307 | 21.0 | 380.23 | 11.98 | 21.0 |
| 1.13081 | 0.0 | 8.14 | 0 | 0.5380 | 5.713 | 94.1 | 4.2330 | 4 | 307 | 21.0 | 360.17 | 22.60 | 12.7 |
| 1.35472 | 0.0 | 8.14 | 0 | 0.5380 | 6.072 | 100.0 | 4.1750 | 4 | 307 | 21.0 | 376.73 | 13.04 | 14.5 |
| 1.38799 | 0.0 | 8.14 | 0 | 0.5380 | 5.950 | 82.0 | 3.9900 | 4 | 307 | 21.0 | 232.60 | 27.71 | 13.2 |
| 1.15172 | 0.0 | 8.14 | 0 | 0.5380 | 5.701 | 95.0 | 3.7872 | 4 | 307 | 21.0 | 358.77 | 18.35 | 13.1 |
| 1.61282 | 0.0 | 8.14 | 0 | 0.5380 | 6.096 | 96.9 | 3.7598 | 4 | 307 | 21.0 | 248.31 | 20.34 | 13.5 |
| 0.06417 | 0.0 | 5.96 | 0 | 0.4990 | 5.933 | 68.2 | 3.3603 | 5 | 279 | 19.2 | 396.90 | 9.68 | 18.9 |
| 0.09744 | 0.0 | 5.96 | 0 | 0.4990 | 5.841 | 61.4 | 3.3779 | 5 | 279 | 19.2 | 377.56 | 11.41 | 20.0 |
| 0.08014 | 0.0 | 5.96 | 0 | 0.4990 | 5.850 | 41.5 | 3.9342 | 5 | 279 | 19.2 | 396.90 | 8.77 | 21.0 |
| 0.17505 | 0.0 | 5.96 | 0 | 0.4990 | 5.966 | 30.2 | 3.8473 | 5 | 279 | 19.2 | 393.43 | 10.13 | 24.7 |
| 0.02763 | 75.0 | 2.95 | 0 | 0.4280 | 6.595 | 21.8 | 5.4011 | 3 | 252 | 18.3 | 395.63 | 4.32 | 30.8 |
| 0.03359 | 75.0 | 2.95 | 0 | 0.4280 | 7.024 | 15.8 | 5.4011 | 3 | 252 | 18.3 | 395.62 | 1.98 | 34.9 |
| 0.12744 | 0.0 | 6.91 | 0 | 0.4480 | 6.770 | 2.9 | 5.7209 | 3 | 233 | 17.9 | 385.41 | 4.84 | 26.6 |
| 0.14150 | 0.0 | 6.91 | 0 | 0.4480 | 6.169 | 6.6 | 5.7209 | 3 | 233 | 17.9 | 383.37 | 5.81 | 25.3 |
| 0.15936 | 0.0 | 6.91 | 0 | 0.4480 | 6.211 | 6.5 | 5.7209 | 3 | 233 | 17.9 | 394.46 | 7.44 | 24.7 |
| 0.12269 | 0.0 | 6.91 | 0 | 0.4480 | 6.069 | 40.0 | 5.7209 | 3 | 233 | 17.9 | 389.39 | 9.55 | 21.2 |
| 0.17142 | 0.0 | 6.91 | 0 | 0.4480 | 5.682 | 33.8 | 5.1004 | 3 | 233 | 17.9 | 396.90 | 10.21 | 19.3 |
| 0.18836 | 0.0 | 6.91 | 0 | 0.4480 | 5.786 | 33.3 | 5.1004 | 3 | 233 | 17.9 | 396.90 | 14.15 | 20.0 |
| 0.22927 | 0.0 | 6.91 | 0 | 0.4480 | 6.030 | 85.5 | 5.6894 | 3 | 233 | 17.9 | 392.74 | 18.80 | 16.6 |
| 0.25387 | 0.0 | 6.91 | 0 | 0.4480 | 5.399 | 95.3 | 5.8700 | 3 | 233 | 17.9 | 396.90 | 30.81 | 14.4 |
| 0.21977 | 0.0 | 6.91 | 0 | 0.4480 | 5.602 | 62.0 | 6.0877 | 3 | 233 | 17.9 | 396.90 | 16.20 | 19.4 |
| 0.08873 | 21.0 | 5.64 | 0 | 0.4390 | 5.963 | 45.7 | 6.8147 | 4 | 243 | 16.8 | 395.56 | 13.45 | 19.7 |
| 0.04337 | 21.0 | 5.64 | 0 | 0.4390 | 6.115 | 63.0 | 6.8147 | 4 | 243 | 16.8 | 393.97 | 9.43 | 20.5 |
| 0.05360 | 21.0 | 5.64 | 0 | 0.4390 | 6.511 | 21.1 | 6.8147 | 4 | 243 | 16.8 | 396.90 | 5.28 | 25.0 |
| 0.04981 | 21.0 | 5.64 | 0 | 0.4390 | 5.998 | 21.4 | 6.8147 | 4 | 243 | 16.8 | 396.90 | 8.43 | 23.4 |
| 0.01360 | 75.0 | 4.00 | 0 | 0.4100 | 5.888 | 47.6 | 7.3197 | 3 | 469 | 21.1 | 396.90 | 14.80 | 18.9 |
| 0.01311 | 90.0 | 1.22 | 0 | 0.4030 | 7.249 | 21.9 | 8.6966 | 5 | 226 | 17.9 | 395.93 | 4.81 | 35.4 |
| 0.02055 | 85.0 | 0.74 | 0 | 0.4100 | 6.383 | 35.7 | 9.1876 | 2 | 313 | 17.3 | 396.90 | 5.77 | 24.7 |
| 0.01432 | 100.0 | 1.32 | 0 | 0.4110 | 6.816 | 40.5 | 8.3248 | 5 | 256 | 15.1 | 392.90 | 3.95 | 31.6 |
| 0.15445 | 25.0 | 5.13 | 0 | 0.4530 | 6.145 | 29.2 | 7.8148 | 8 | 284 | 19.7 | 390.68 | 6.86 | 23.3 |
| 0.10328 | 25.0 | 5.13 | 0 | 0.4530 | 5.927 | 47.2 | 6.9320 | 8 | 284 | 19.7 | 396.90 | 9.22 | 19.6 |
| 0.14932 | 25.0 | 5.13 | 0 | 0.4530 | 5.741 | 66.2 | 7.2254 | 8 | 284 | 19.7 | 395.11 | 13.15 | 18.7 |
| 0.17171 | 25.0 | 5.13 | 0 | 0.4530 | 5.966 | 93.4 | 6.8185 | 8 | 284 | 19.7 | 378.08 | 14.44 | 16.0 |
| 0.11027 | 25.0 | 5.13 | 0 | 0.4530 | 6.456 | 67.8 | 7.2255 | 8 | 284 | 19.7 | 396.90 | 6.73 | 22.2 |
| 0.12650 | 25.0 | 5.13 | 0 | 0.4530 | 6.762 | 43.4 | 7.9809 | 8 | 284 | 19.7 | 395.58 | 9.50 | 25.0 |
| 0.01951 | 17.5 | 1.38 | 0 | 0.4161 | 7.104 | 59.5 | 9.2229 | 3 | 216 | 18.6 | 393.24 | 8.05 | 33.0 |
| 0.03584 | 80.0 | 3.37 | 0 | 0.3980 | 6.290 | 17.8 | 6.6115 | 4 | 337 | 16.1 | 396.90 | 4.67 | 23.5 |
| 0.04379 | 80.0 | 3.37 | 0 | 0.3980 | 5.787 | 31.1 | 6.6115 | 4 | 337 | 16.1 | 396.90 | 10.24 | 19.4 |
| 0.05789 | 12.5 | 6.07 | 0 | 0.4090 | 5.878 | 21.4 | 6.4980 | 4 | 345 | 18.9 | 396.21 | 8.10 | 22.0 |
| 0.13554 | 12.5 | 6.07 | 0 | 0.4090 | 5.594 | 36.8 | 6.4980 | 4 | 345 | 18.9 | 396.90 | 13.09 | 17.4 |
| 0.12816 | 12.5 | 6.07 | 0 | 0.4090 | 5.885 | 33.0 | 6.4980 | 4 | 345 | 18.9 | 396.90 | 8.79 | 20.9 |
| 0.08826 | 0.0 | 10.81 | 0 | 0.4130 | 6.417 | 6.6 | 5.2873 | 4 | 305 | 19.2 | 383.73 | 6.72 | 24.2 |
| 0.15876 | 0.0 | 10.81 | 0 | 0.4130 | 5.961 | 17.5 | 5.2873 | 4 | 305 | 19.2 | 376.94 | 9.88 | 21.7 |
| 0.09164 | 0.0 | 10.81 | 0 | 0.4130 | 6.065 | 7.8 | 5.2873 | 4 | 305 | 19.2 | 390.91 | 5.52 | 22.8 |
| 0.19539 | 0.0 | 10.81 | 0 | 0.4130 | 6.245 | 6.2 | 5.2873 | 4 | 305 | 19.2 | 377.17 | 7.54 | 23.4 |
| 0.07896 | 0.0 | 12.83 | 0 | 0.4370 | 6.273 | 6.0 | 4.2515 | 5 | 398 | 18.7 | 394.92 | 6.78 | 24.1 |
| 0.09512 | 0.0 | 12.83 | 0 | 0.4370 | 6.286 | 45.0 | 4.5026 | 5 | 398 | 18.7 | 383.23 | 8.94 | 21.4 |
| 0.10153 | 0.0 | 12.83 | 0 | 0.4370 | 6.279 | 74.5 | 4.0522 | 5 | 398 | 18.7 | 373.66 | 11.97 | 20.0 |
| 0.08707 | 0.0 | 12.83 | 0 | 0.4370 | 6.140 | 45.8 | 4.0905 | 5 | 398 | 18.7 | 386.96 | 10.27 | 20.8 |
| 0.05646 | 0.0 | 12.83 | 0 | 0.4370 | 6.232 | 53.7 | 5.0141 | 5 | 398 | 18.7 | 386.40 | 12.34 | 21.2 |
| 0.08387 | 0.0 | 12.83 | 0 | 0.4370 | 5.874 | 36.6 | 4.5026 | 5 | 398 | 18.7 | 396.06 | 9.10 | 20.3 |
| 0.04113 | 25.0 | 4.86 | 0 | 0.4260 | 6.727 | 33.5 | 5.4007 | 4 | 281 | 19.0 | 396.90 | 5.29 | 28.0 |
| 0.04462 | 25.0 | 4.86 | 0 | 0.4260 | 6.619 | 70.4 | 5.4007 | 4 | 281 | 19.0 | 395.63 | 7.22 | 23.9 |
| 0.03659 | 25.0 | 4.86 | 0 | 0.4260 | 6.302 | 32.2 | 5.4007 | 4 | 281 | 19.0 | 396.90 | 6.72 | 24.8 |
| 0.03551 | 25.0 | 4.86 | 0 | 0.4260 | 6.167 | 46.7 | 5.4007 | 4 | 281 | 19.0 | 390.64 | 7.51 | 22.9 |
| 0.05059 | 0.0 | 4.49 | 0 | 0.4490 | 6.389 | 48.0 | 4.7794 | 3 | 247 | 18.5 | 396.90 | 9.62 | 23.9 |
| 0.05735 | 0.0 | 4.49 | 0 | 0.4490 | 6.630 | 56.1 | 4.4377 | 3 | 247 | 18.5 | 392.30 | 6.53 | 26.6 |
| 0.05188 | 0.0 | 4.49 | 0 | 0.4490 | 6.015 | 45.1 | 4.4272 | 3 | 247 | 18.5 | 395.99 | 12.86 | 22.5 |
| 0.07151 | 0.0 | 4.49 | 0 | 0.4490 | 6.121 | 56.8 | 3.7476 | 3 | 247 | 18.5 | 395.15 | 8.44 | 22.2 |
| 0.05660 | 0.0 | 3.41 | 0 | 0.4890 | 7.007 | 86.3 | 3.4217 | 2 | 270 | 17.8 | 396.90 | 5.50 | 23.6 |
| 0.05302 | 0.0 | 3.41 | 0 | 0.4890 | 7.079 | 63.1 | 3.4145 | 2 | 270 | 17.8 | 396.06 | 5.70 | 28.7 |
| 0.04684 | 0.0 | 3.41 | 0 | 0.4890 | 6.417 | 66.1 | 3.0923 | 2 | 270 | 17.8 | 392.18 | 8.81 | 22.6 |
| 0.03932 | 0.0 | 3.41 | 0 | 0.4890 | 6.405 | 73.9 | 3.0921 | 2 | 270 | 17.8 | 393.55 | 8.20 | 22.0 |
| 0.04203 | 28.0 | 15.04 | 0 | 0.4640 | 6.442 | 53.6 | 3.6659 | 4 | 270 | 18.2 | 395.01 | 8.16 | 22.9 |
| 0.02875 | 28.0 | 15.04 | 0 | 0.4640 | 6.211 | 28.9 | 3.6659 | 4 | 270 | 18.2 | 396.33 | 6.21 | 25.0 |
| 0.04294 | 28.0 | 15.04 | 0 | 0.4640 | 6.249 | 77.3 | 3.6150 | 4 | 270 | 18.2 | 396.90 | 10.59 | 20.6 |
| 0.12204 | 0.0 | 2.89 | 0 | 0.4450 | 6.625 | 57.8 | 3.4952 | 2 | 276 | 18.0 | 357.98 | 6.65 | 28.4 |
| 0.11504 | 0.0 | 2.89 | 0 | 0.4450 | 6.163 | 69.6 | 3.4952 | 2 | 276 | 18.0 | 391.83 | 11.34 | 21.4 |
| 0.12083 | 0.0 | 2.89 | 0 | 0.4450 | 8.069 | 76.0 | 3.4952 | 2 | 276 | 18.0 | 396.90 | 4.21 | 38.7 |
| 0.08187 | 0.0 | 2.89 | 0 | 0.4450 | 7.820 | 36.9 | 3.4952 | 2 | 276 | 18.0 | 393.53 | 3.57 | 43.8 |
| 0.06860 | 0.0 | 2.89 | 0 | 0.4450 | 7.416 | 62.5 | 3.4952 | 2 | 276 | 18.0 | 396.90 | 6.19 | 33.2 |
| 0.14866 | 0.0 | 8.56 | 0 | 0.5200 | 6.727 | 79.9 | 2.7778 | 5 | 384 | 20.9 | 394.76 | 9.42 | 27.5 |
| 0.11432 | 0.0 | 8.56 | 0 | 0.5200 | 6.781 | 71.3 | 2.8561 | 5 | 384 | 20.9 | 395.58 | 7.67 | 26.5 |
| 0.22876 | 0.0 | 8.56 | 0 | 0.5200 | 6.405 | 85.4 | 2.7147 | 5 | 384 | 20.9 | 70.80 | 10.63 | 18.6 |
| 0.21161 | 0.0 | 8.56 | 0 | 0.5200 | 6.137 | 87.4 | 2.7147 | 5 | 384 | 20.9 | 394.47 | 13.44 | 19.3 |
| 0.13960 | 0.0 | 8.56 | 0 | 0.5200 | 6.167 | 90.0 | 2.4210 | 5 | 384 | 20.9 | 392.69 | 12.33 | 20.1 |
| 0.13262 | 0.0 | 8.56 | 0 | 0.5200 | 5.851 | 96.7 | 2.1069 | 5 | 384 | 20.9 | 394.05 | 16.47 | 19.5 |
| 0.17120 | 0.0 | 8.56 | 0 | 0.5200 | 5.836 | 91.9 | 2.2110 | 5 | 384 | 20.9 | 395.67 | 18.66 | 19.5 |
| 0.13117 | 0.0 | 8.56 | 0 | 0.5200 | 6.127 | 85.2 | 2.1224 | 5 | 384 | 20.9 | 387.69 | 14.09 | 20.4 |
| 0.12802 | 0.0 | 8.56 | 0 | 0.5200 | 6.474 | 97.1 | 2.4329 | 5 | 384 | 20.9 | 395.24 | 12.27 | 19.8 |
| 0.26363 | 0.0 | 8.56 | 0 | 0.5200 | 6.229 | 91.2 | 2.5451 | 5 | 384 | 20.9 | 391.23 | 15.55 | 19.4 |
| 0.10793 | 0.0 | 8.56 | 0 | 0.5200 | 6.195 | 54.4 | 2.7778 | 5 | 384 | 20.9 | 393.49 | 13.00 | 21.7 |
| 0.10084 | 0.0 | 10.01 | 0 | 0.5470 | 6.715 | 81.6 | 2.6775 | 6 | 432 | 17.8 | 395.59 | 10.16 | 22.8 |
| 0.12329 | 0.0 | 10.01 | 0 | 0.5470 | 5.913 | 92.9 | 2.3534 | 6 | 432 | 17.8 | 394.95 | 16.21 | 18.8 |
| 0.22212 | 0.0 | 10.01 | 0 | 0.5470 | 6.092 | 95.4 | 2.5480 | 6 | 432 | 17.8 | 396.90 | 17.09 | 18.7 |
| 0.14231 | 0.0 | 10.01 | 0 | 0.5470 | 6.254 | 84.2 | 2.2565 | 6 | 432 | 17.8 | 388.74 | 10.45 | 18.5 |
| 0.17134 | 0.0 | 10.01 | 0 | 0.5470 | 5.928 | 88.2 | 2.4631 | 6 | 432 | 17.8 | 344.91 | 15.76 | 18.3 |
| 0.13158 | 0.0 | 10.01 | 0 | 0.5470 | 6.176 | 72.5 | 2.7301 | 6 | 432 | 17.8 | 393.30 | 12.04 | 21.2 |
| 0.15098 | 0.0 | 10.01 | 0 | 0.5470 | 6.021 | 82.6 | 2.7474 | 6 | 432 | 17.8 | 394.51 | 10.30 | 19.2 |
| 0.13058 | 0.0 | 10.01 | 0 | 0.5470 | 5.872 | 73.1 | 2.4775 | 6 | 432 | 17.8 | 338.63 | 15.37 | 20.4 |
| 0.14476 | 0.0 | 10.01 | 0 | 0.5470 | 5.731 | 65.2 | 2.7592 | 6 | 432 | 17.8 | 391.50 | 13.61 | 19.3 |
| 0.06899 | 0.0 | 25.65 | 0 | 0.5810 | 5.870 | 69.7 | 2.2577 | 2 | 188 | 19.1 | 389.15 | 14.37 | 22.0 |
| 0.07165 | 0.0 | 25.65 | 0 | 0.5810 | 6.004 | 84.1 | 2.1974 | 2 | 188 | 19.1 | 377.67 | 14.27 | 20.3 |
| 0.09299 | 0.0 | 25.65 | 0 | 0.5810 | 5.961 | 92.9 | 2.0869 | 2 | 188 | 19.1 | 378.09 | 17.93 | 20.5 |
| 0.15038 | 0.0 | 25.65 | 0 | 0.5810 | 5.856 | 97.0 | 1.9444 | 2 | 188 | 19.1 | 370.31 | 25.41 | 17.3 |
| 0.09849 | 0.0 | 25.65 | 0 | 0.5810 | 5.879 | 95.8 | 2.0063 | 2 | 188 | 19.1 | 379.38 | 17.58 | 18.8 |
| 0.16902 | 0.0 | 25.65 | 0 | 0.5810 | 5.986 | 88.4 | 1.9929 | 2 | 188 | 19.1 | 385.02 | 14.81 | 21.4 |
| 0.38735 | 0.0 | 25.65 | 0 | 0.5810 | 5.613 | 95.6 | 1.7572 | 2 | 188 | 19.1 | 359.29 | 27.26 | 15.7 |
| 0.25915 | 0.0 | 21.89 | 0 | 0.6240 | 5.693 | 96.0 | 1.7883 | 4 | 437 | 21.2 | 392.11 | 17.19 | 16.2 |
| 0.32543 | 0.0 | 21.89 | 0 | 0.6240 | 6.431 | 98.8 | 1.8125 | 4 | 437 | 21.2 | 396.90 | 15.39 | 18.0 |
| 0.88125 | 0.0 | 21.89 | 0 | 0.6240 | 5.637 | 94.7 | 1.9799 | 4 | 437 | 21.2 | 396.90 | 18.34 | 14.3 |
| 0.34006 | 0.0 | 21.89 | 0 | 0.6240 | 6.458 | 98.9 | 2.1185 | 4 | 437 | 21.2 | 395.04 | 12.60 | 19.2 |
| 1.19294 | 0.0 | 21.89 | 0 | 0.6240 | 6.326 | 97.7 | 2.2710 | 4 | 437 | 21.2 | 396.90 | 12.26 | 19.6 |
| 0.59005 | 0.0 | 21.89 | 0 | 0.6240 | 6.372 | 97.9 | 2.3274 | 4 | 437 | 21.2 | 385.76 | 11.12 | 23.0 |
| 0.32982 | 0.0 | 21.89 | 0 | 0.6240 | 5.822 | 95.4 | 2.4699 | 4 | 437 | 21.2 | 388.69 | 15.03 | 18.4 |
| 0.97617 | 0.0 | 21.89 | 0 | 0.6240 | 5.757 | 98.4 | 2.3460 | 4 | 437 | 21.2 | 262.76 | 17.31 | 15.6 |
| 0.55778 | 0.0 | 21.89 | 0 | 0.6240 | 6.335 | 98.2 | 2.1107 | 4 | 437 | 21.2 | 394.67 | 16.96 | 18.1 |
| 0.32264 | 0.0 | 21.89 | 0 | 0.6240 | 5.942 | 93.5 | 1.9669 | 4 | 437 | 21.2 | 378.25 | 16.90 | 17.4 |
| 0.35233 | 0.0 | 21.89 | 0 | 0.6240 | 6.454 | 98.4 | 1.8498 | 4 | 437 | 21.2 | 394.08 | 14.59 | 17.1 |
| 0.24980 | 0.0 | 21.89 | 0 | 0.6240 | 5.857 | 98.2 | 1.6686 | 4 | 437 | 21.2 | 392.04 | 21.32 | 13.3 |
| 0.54452 | 0.0 | 21.89 | 0 | 0.6240 | 6.151 | 97.9 | 1.6687 | 4 | 437 | 21.2 | 396.90 | 18.46 | 17.8 |
| 0.29090 | 0.0 | 21.89 | 0 | 0.6240 | 6.174 | 93.6 | 1.6119 | 4 | 437 | 21.2 | 388.08 | 24.16 | 14.0 |
| 1.62864 | 0.0 | 21.89 | 0 | 0.6240 | 5.019 | 100.0 | 1.4394 | 4 | 437 | 21.2 | 396.90 | 34.41 | 14.4 |
| 3.32105 | 0.0 | 19.58 | 1 | 0.8710 | 5.403 | 100.0 | 1.3216 | 5 | 403 | 14.7 | 396.90 | 26.82 | 13.4 |
| 4.09740 | 0.0 | 19.58 | 0 | 0.8710 | 5.468 | 100.0 | 1.4118 | 5 | 403 | 14.7 | 396.90 | 26.42 | 15.6 |
| 2.77974 | 0.0 | 19.58 | 0 | 0.8710 | 4.903 | 97.8 | 1.3459 | 5 | 403 | 14.7 | 396.90 | 29.29 | 11.8 |
| 2.37934 | 0.0 | 19.58 | 0 | 0.8710 | 6.130 | 100.0 | 1.4191 | 5 | 403 | 14.7 | 172.91 | 27.80 | 13.8 |
| 2.15505 | 0.0 | 19.58 | 0 | 0.8710 | 5.628 | 100.0 | 1.5166 | 5 | 403 | 14.7 | 169.27 | 16.65 | 15.6 |
| 2.36862 | 0.0 | 19.58 | 0 | 0.8710 | 4.926 | 95.7 | 1.4608 | 5 | 403 | 14.7 | 391.71 | 29.53 | 14.6 |
| 2.33099 | 0.0 | 19.58 | 0 | 0.8710 | 5.186 | 93.8 | 1.5296 | 5 | 403 | 14.7 | 356.99 | 28.32 | 17.8 |
| 2.73397 | 0.0 | 19.58 | 0 | 0.8710 | 5.597 | 94.9 | 1.5257 | 5 | 403 | 14.7 | 351.85 | 21.45 | 15.4 |
| 1.65660 | 0.0 | 19.58 | 0 | 0.8710 | 6.122 | 97.3 | 1.6180 | 5 | 403 | 14.7 | 372.80 | 14.10 | 21.5 |
| 1.49632 | 0.0 | 19.58 | 0 | 0.8710 | 5.404 | 100.0 | 1.5916 | 5 | 403 | 14.7 | 341.60 | 13.28 | 19.6 |
| 1.12658 | 0.0 | 19.58 | 1 | 0.8710 | 5.012 | 88.0 | 1.6102 | 5 | 403 | 14.7 | 343.28 | 12.12 | 15.3 |
| 2.14918 | 0.0 | 19.58 | 0 | 0.8710 | 5.709 | 98.5 | 1.6232 | 5 | 403 | 14.7 | 261.95 | 15.79 | 19.4 |
| 1.41385 | 0.0 | 19.58 | 1 | 0.8710 | 6.129 | 96.0 | 1.7494 | 5 | 403 | 14.7 | 321.02 | 15.12 | 17.0 |
| 3.53501 | 0.0 | 19.58 | 1 | 0.8710 | 6.152 | 82.6 | 1.7455 | 5 | 403 | 14.7 | 88.01 | 15.02 | 15.6 |
| 2.44668 | 0.0 | 19.58 | 0 | 0.8710 | 5.272 | 94.0 | 1.7364 | 5 | 403 | 14.7 | 88.63 | 16.14 | 13.1 |
| 1.22358 | 0.0 | 19.58 | 0 | 0.6050 | 6.943 | 97.4 | 1.8773 | 5 | 403 | 14.7 | 363.43 | 4.59 | 41.3 |
| 1.34284 | 0.0 | 19.58 | 0 | 0.6050 | 6.066 | 100.0 | 1.7573 | 5 | 403 | 14.7 | 353.89 | 6.43 | 24.3 |
| 1.42502 | 0.0 | 19.58 | 0 | 0.8710 | 6.510 | 100.0 | 1.7659 | 5 | 403 | 14.7 | 364.31 | 7.39 | 23.3 |
| 1.27346 | 0.0 | 19.58 | 1 | 0.6050 | 6.250 | 92.6 | 1.7984 | 5 | 403 | 14.7 | 338.92 | 5.50 | 27.0 |
| 1.46336 | 0.0 | 19.58 | 0 | 0.6050 | 7.489 | 90.8 | 1.9709 | 5 | 403 | 14.7 | 374.43 | 1.73 | 50.0 |
| 1.83377 | 0.0 | 19.58 | 1 | 0.6050 | 7.802 | 98.2 | 2.0407 | 5 | 403 | 14.7 | 389.61 | 1.92 | 50.0 |
| 1.51902 | 0.0 | 19.58 | 1 | 0.6050 | 8.375 | 93.9 | 2.1620 | 5 | 403 | 14.7 | 388.45 | 3.32 | 50.0 |
| 2.24236 | 0.0 | 19.58 | 0 | 0.6050 | 5.854 | 91.8 | 2.4220 | 5 | 403 | 14.7 | 395.11 | 11.64 | 22.7 |
| 2.92400 | 0.0 | 19.58 | 0 | 0.6050 | 6.101 | 93.0 | 2.2834 | 5 | 403 | 14.7 | 240.16 | 9.81 | 25.0 |
| 2.01019 | 0.0 | 19.58 | 0 | 0.6050 | 7.929 | 96.2 | 2.0459 | 5 | 403 | 14.7 | 369.30 | 3.70 | 50.0 |
| 1.80028 | 0.0 | 19.58 | 0 | 0.6050 | 5.877 | 79.2 | 2.4259 | 5 | 403 | 14.7 | 227.61 | 12.14 | 23.8 |
| 2.30040 | 0.0 | 19.58 | 0 | 0.6050 | 6.319 | 96.1 | 2.1000 | 5 | 403 | 14.7 | 297.09 | 11.10 | 23.8 |
| 2.44953 | 0.0 | 19.58 | 0 | 0.6050 | 6.402 | 95.2 | 2.2625 | 5 | 403 | 14.7 | 330.04 | 11.32 | 22.3 |
| 1.20742 | 0.0 | 19.58 | 0 | 0.6050 | 5.875 | 94.6 | 2.4259 | 5 | 403 | 14.7 | 292.29 | 14.43 | 17.4 |
| 2.31390 | 0.0 | 19.58 | 0 | 0.6050 | 5.880 | 97.3 | 2.3887 | 5 | 403 | 14.7 | 348.13 | 12.03 | 19.1 |
| 0.13914 | 0.0 | 4.05 | 0 | 0.5100 | 5.572 | 88.5 | 2.5961 | 5 | 296 | 16.6 | 396.90 | 14.69 | 23.1 |
| 0.09178 | 0.0 | 4.05 | 0 | 0.5100 | 6.416 | 84.1 | 2.6463 | 5 | 296 | 16.6 | 395.50 | 9.04 | 23.6 |
| 0.08447 | 0.0 | 4.05 | 0 | 0.5100 | 5.859 | 68.7 | 2.7019 | 5 | 296 | 16.6 | 393.23 | 9.64 | 22.6 |
| 0.06664 | 0.0 | 4.05 | 0 | 0.5100 | 6.546 | 33.1 | 3.1323 | 5 | 296 | 16.6 | 390.96 | 5.33 | 29.4 |
| 0.07022 | 0.0 | 4.05 | 0 | 0.5100 | 6.020 | 47.2 | 3.5549 | 5 | 296 | 16.6 | 393.23 | 10.11 | 23.2 |
| 0.05425 | 0.0 | 4.05 | 0 | 0.5100 | 6.315 | 73.4 | 3.3175 | 5 | 296 | 16.6 | 395.60 | 6.29 | 24.6 |
| 0.06642 | 0.0 | 4.05 | 0 | 0.5100 | 6.860 | 74.4 | 2.9153 | 5 | 296 | 16.6 | 391.27 | 6.92 | 29.9 |
| 0.05780 | 0.0 | 2.46 | 0 | 0.4880 | 6.980 | 58.4 | 2.8290 | 3 | 193 | 17.8 | 396.90 | 5.04 | 37.2 |
| 0.06588 | 0.0 | 2.46 | 0 | 0.4880 | 7.765 | 83.3 | 2.7410 | 3 | 193 | 17.8 | 395.56 | 7.56 | 39.8 |
| 0.06888 | 0.0 | 2.46 | 0 | 0.4880 | 6.144 | 62.2 | 2.5979 | 3 | 193 | 17.8 | 396.90 | 9.45 | 36.2 |
| 0.09103 | 0.0 | 2.46 | 0 | 0.4880 | 7.155 | 92.2 | 2.7006 | 3 | 193 | 17.8 | 394.12 | 4.82 | 37.9 |
| 0.10008 | 0.0 | 2.46 | 0 | 0.4880 | 6.563 | 95.6 | 2.8470 | 3 | 193 | 17.8 | 396.90 | 5.68 | 32.5 |
| 0.08308 | 0.0 | 2.46 | 0 | 0.4880 | 5.604 | 89.8 | 2.9879 | 3 | 193 | 17.8 | 391.00 | 13.98 | 26.4 |
| 0.06047 | 0.0 | 2.46 | 0 | 0.4880 | 6.153 | 68.8 | 3.2797 | 3 | 193 | 17.8 | 387.11 | 13.15 | 29.6 |
| 0.05602 | 0.0 | 2.46 | 0 | 0.4880 | 7.831 | 53.6 | 3.1992 | 3 | 193 | 17.8 | 392.63 | 4.45 | 50.0 |
| 0.07875 | 45.0 | 3.44 | 0 | 0.4370 | 6.782 | 41.1 | 3.7886 | 5 | 398 | 15.2 | 393.87 | 6.68 | 32.0 |
| 0.12579 | 45.0 | 3.44 | 0 | 0.4370 | 6.556 | 29.1 | 4.5667 | 5 | 398 | 15.2 | 382.84 | 4.56 | 29.8 |
| 0.08370 | 45.0 | 3.44 | 0 | 0.4370 | 7.185 | 38.9 | 4.5667 | 5 | 398 | 15.2 | 396.90 | 5.39 | 34.9 |
| 0.09068 | 45.0 | 3.44 | 0 | 0.4370 | 6.951 | 21.5 | 6.4798 | 5 | 398 | 15.2 | 377.68 | 5.10 | 37.0 |
| 0.06911 | 45.0 | 3.44 | 0 | 0.4370 | 6.739 | 30.8 | 6.4798 | 5 | 398 | 15.2 | 389.71 | 4.69 | 30.5 |
| 0.08664 | 45.0 | 3.44 | 0 | 0.4370 | 7.178 | 26.3 | 6.4798 | 5 | 398 | 15.2 | 390.49 | 2.87 | 36.4 |
| 0.02187 | 60.0 | 2.93 | 0 | 0.4010 | 6.800 | 9.9 | 6.2196 | 1 | 265 | 15.6 | 393.37 | 5.03 | 31.1 |
| 0.01439 | 60.0 | 2.93 | 0 | 0.4010 | 6.604 | 18.8 | 6.2196 | 1 | 265 | 15.6 | 376.70 | 4.38 | 29.1 |
| 0.01381 | 80.0 | 0.46 | 0 | 0.4220 | 7.875 | 32.0 | 5.6484 | 4 | 255 | 14.4 | 394.23 | 2.97 | 50.0 |
| 0.04011 | 80.0 | 1.52 | 0 | 0.4040 | 7.287 | 34.1 | 7.3090 | 2 | 329 | 12.6 | 396.90 | 4.08 | 33.3 |
| 0.04666 | 80.0 | 1.52 | 0 | 0.4040 | 7.107 | 36.6 | 7.3090 | 2 | 329 | 12.6 | 354.31 | 8.61 | 30.3 |
| 0.03768 | 80.0 | 1.52 | 0 | 0.4040 | 7.274 | 38.3 | 7.3090 | 2 | 329 | 12.6 | 392.20 | 6.62 | 34.6 |
| 0.03150 | 95.0 | 1.47 | 0 | 0.4030 | 6.975 | 15.3 | 7.6534 | 3 | 402 | 17.0 | 396.90 | 4.56 | 34.9 |
| 0.01778 | 95.0 | 1.47 | 0 | 0.4030 | 7.135 | 13.9 | 7.6534 | 3 | 402 | 17.0 | 384.30 | 4.45 | 32.9 |
| 0.03445 | 82.5 | 2.03 | 0 | 0.4150 | 6.162 | 38.4 | 6.2700 | 2 | 348 | 14.7 | 393.77 | 7.43 | 24.1 |
| 0.02177 | 82.5 | 2.03 | 0 | 0.4150 | 7.610 | 15.7 | 6.2700 | 2 | 348 | 14.7 | 395.38 | 3.11 | 42.3 |
| 0.03510 | 95.0 | 2.68 | 0 | 0.4161 | 7.853 | 33.2 | 5.1180 | 4 | 224 | 14.7 | 392.78 | 3.81 | 48.5 |
| 0.02009 | 95.0 | 2.68 | 0 | 0.4161 | 8.034 | 31.9 | 5.1180 | 4 | 224 | 14.7 | 390.55 | 2.88 | 50.0 |
| 0.13642 | 0.0 | 10.59 | 0 | 0.4890 | 5.891 | 22.3 | 3.9454 | 4 | 277 | 18.6 | 396.90 | 10.87 | 22.6 |
| 0.22969 | 0.0 | 10.59 | 0 | 0.4890 | 6.326 | 52.5 | 4.3549 | 4 | 277 | 18.6 | 394.87 | 10.97 | 24.4 |
| 0.25199 | 0.0 | 10.59 | 0 | 0.4890 | 5.783 | 72.7 | 4.3549 | 4 | 277 | 18.6 | 389.43 | 18.06 | 22.5 |
| 0.13587 | 0.0 | 10.59 | 1 | 0.4890 | 6.064 | 59.1 | 4.2392 | 4 | 277 | 18.6 | 381.32 | 14.66 | 24.4 |
| 0.43571 | 0.0 | 10.59 | 1 | 0.4890 | 5.344 | 100.0 | 3.8750 | 4 | 277 | 18.6 | 396.90 | 23.09 | 20.0 |
| 0.17446 | 0.0 | 10.59 | 1 | 0.4890 | 5.960 | 92.1 | 3.8771 | 4 | 277 | 18.6 | 393.25 | 17.27 | 21.7 |
| 0.37578 | 0.0 | 10.59 | 1 | 0.4890 | 5.404 | 88.6 | 3.6650 | 4 | 277 | 18.6 | 395.24 | 23.98 | 19.3 |
| 0.21719 | 0.0 | 10.59 | 1 | 0.4890 | 5.807 | 53.8 | 3.6526 | 4 | 277 | 18.6 | 390.94 | 16.03 | 22.4 |
| 0.14052 | 0.0 | 10.59 | 0 | 0.4890 | 6.375 | 32.3 | 3.9454 | 4 | 277 | 18.6 | 385.81 | 9.38 | 28.1 |
| 0.28955 | 0.0 | 10.59 | 0 | 0.4890 | 5.412 | 9.8 | 3.5875 | 4 | 277 | 18.6 | 348.93 | 29.55 | 23.7 |
| 0.19802 | 0.0 | 10.59 | 0 | 0.4890 | 6.182 | 42.4 | 3.9454 | 4 | 277 | 18.6 | 393.63 | 9.47 | 25.0 |
| 0.04560 | 0.0 | 13.89 | 1 | 0.5500 | 5.888 | 56.0 | 3.1121 | 5 | 276 | 16.4 | 392.80 | 13.51 | 23.3 |
| 0.07013 | 0.0 | 13.89 | 0 | 0.5500 | 6.642 | 85.1 | 3.4211 | 5 | 276 | 16.4 | 392.78 | 9.69 | 28.7 |
| 0.11069 | 0.0 | 13.89 | 1 | 0.5500 | 5.951 | 93.8 | 2.8893 | 5 | 276 | 16.4 | 396.90 | 17.92 | 21.5 |
| 0.11425 | 0.0 | 13.89 | 1 | 0.5500 | 6.373 | 92.4 | 3.3633 | 5 | 276 | 16.4 | 393.74 | 10.50 | 23.0 |
| 0.35809 | 0.0 | 6.20 | 1 | 0.5070 | 6.951 | 88.5 | 2.8617 | 8 | 307 | 17.4 | 391.70 | 9.71 | 26.7 |
| 0.40771 | 0.0 | 6.20 | 1 | 0.5070 | 6.164 | 91.3 | 3.0480 | 8 | 307 | 17.4 | 395.24 | 21.46 | 21.7 |
| 0.62356 | 0.0 | 6.20 | 1 | 0.5070 | 6.879 | 77.7 | 3.2721 | 8 | 307 | 17.4 | 390.39 | 9.93 | 27.5 |
| 0.61470 | 0.0 | 6.20 | 0 | 0.5070 | 6.618 | 80.8 | 3.2721 | 8 | 307 | 17.4 | 396.90 | 7.60 | 30.1 |
| 0.31533 | 0.0 | 6.20 | 0 | 0.5040 | 8.266 | 78.3 | 2.8944 | 8 | 307 | 17.4 | 385.05 | 4.14 | 44.8 |
| 0.52693 | 0.0 | 6.20 | 0 | 0.5040 | 8.725 | 83.0 | 2.8944 | 8 | 307 | 17.4 | 382.00 | 4.63 | 50.0 |
| 0.38214 | 0.0 | 6.20 | 0 | 0.5040 | 8.040 | 86.5 | 3.2157 | 8 | 307 | 17.4 | 387.38 | 3.13 | 37.6 |
| 0.41238 | 0.0 | 6.20 | 0 | 0.5040 | 7.163 | 79.9 | 3.2157 | 8 | 307 | 17.4 | 372.08 | 6.36 | 31.6 |
| 0.29819 | 0.0 | 6.20 | 0 | 0.5040 | 7.686 | 17.0 | 3.3751 | 8 | 307 | 17.4 | 377.51 | 3.92 | 46.7 |
| 0.44178 | 0.0 | 6.20 | 0 | 0.5040 | 6.552 | 21.4 | 3.3751 | 8 | 307 | 17.4 | 380.34 | 3.76 | 31.5 |
| 0.53700 | 0.0 | 6.20 | 0 | 0.5040 | 5.981 | 68.1 | 3.6715 | 8 | 307 | 17.4 | 378.35 | 11.65 | 24.3 |
| 0.46296 | 0.0 | 6.20 | 0 | 0.5040 | 7.412 | 76.9 | 3.6715 | 8 | 307 | 17.4 | 376.14 | 5.25 | 31.7 |
| 0.57529 | 0.0 | 6.20 | 0 | 0.5070 | 8.337 | 73.3 | 3.8384 | 8 | 307 | 17.4 | 385.91 | 2.47 | 41.7 |
| 0.33147 | 0.0 | 6.20 | 0 | 0.5070 | 8.247 | 70.4 | 3.6519 | 8 | 307 | 17.4 | 378.95 | 3.95 | 48.3 |
| 0.44791 | 0.0 | 6.20 | 1 | 0.5070 | 6.726 | 66.5 | 3.6519 | 8 | 307 | 17.4 | 360.20 | 8.05 | 29.0 |
| 0.33045 | 0.0 | 6.20 | 0 | 0.5070 | 6.086 | 61.5 | 3.6519 | 8 | 307 | 17.4 | 376.75 | 10.88 | 24.0 |
| 0.52058 | 0.0 | 6.20 | 1 | 0.5070 | 6.631 | 76.5 | 4.1480 | 8 | 307 | 17.4 | 388.45 | 9.54 | 25.1 |
| 0.51183 | 0.0 | 6.20 | 0 | 0.5070 | 7.358 | 71.6 | 4.1480 | 8 | 307 | 17.4 | 390.07 | 4.73 | 31.5 |
| 0.08244 | 30.0 | 4.93 | 0 | 0.4280 | 6.481 | 18.5 | 6.1899 | 6 | 300 | 16.6 | 379.41 | 6.36 | 23.7 |
| 0.09252 | 30.0 | 4.93 | 0 | 0.4280 | 6.606 | 42.2 | 6.1899 | 6 | 300 | 16.6 | 383.78 | 7.37 | 23.3 |
| 0.11329 | 30.0 | 4.93 | 0 | 0.4280 | 6.897 | 54.3 | 6.3361 | 6 | 300 | 16.6 | 391.25 | 11.38 | 22.0 |
| 0.10612 | 30.0 | 4.93 | 0 | 0.4280 | 6.095 | 65.1 | 6.3361 | 6 | 300 | 16.6 | 394.62 | 12.40 | 20.1 |
| 0.10290 | 30.0 | 4.93 | 0 | 0.4280 | 6.358 | 52.9 | 7.0355 | 6 | 300 | 16.6 | 372.75 | 11.22 | 22.2 |
| 0.12757 | 30.0 | 4.93 | 0 | 0.4280 | 6.393 | 7.8 | 7.0355 | 6 | 300 | 16.6 | 374.71 | 5.19 | 23.7 |
| 0.20608 | 22.0 | 5.86 | 0 | 0.4310 | 5.593 | 76.5 | 7.9549 | 7 | 330 | 19.1 | 372.49 | 12.50 | 17.6 |
| 0.19133 | 22.0 | 5.86 | 0 | 0.4310 | 5.605 | 70.2 | 7.9549 | 7 | 330 | 19.1 | 389.13 | 18.46 | 18.5 |
| 0.33983 | 22.0 | 5.86 | 0 | 0.4310 | 6.108 | 34.9 | 8.0555 | 7 | 330 | 19.1 | 390.18 | 9.16 | 24.3 |
| 0.19657 | 22.0 | 5.86 | 0 | 0.4310 | 6.226 | 79.2 | 8.0555 | 7 | 330 | 19.1 | 376.14 | 10.15 | 20.5 |
| 0.16439 | 22.0 | 5.86 | 0 | 0.4310 | 6.433 | 49.1 | 7.8265 | 7 | 330 | 19.1 | 374.71 | 9.52 | 24.5 |
| 0.19073 | 22.0 | 5.86 | 0 | 0.4310 | 6.718 | 17.5 | 7.8265 | 7 | 330 | 19.1 | 393.74 | 6.56 | 26.2 |
| 0.14030 | 22.0 | 5.86 | 0 | 0.4310 | 6.487 | 13.0 | 7.3967 | 7 | 330 | 19.1 | 396.28 | 5.90 | 24.4 |
| 0.21409 | 22.0 | 5.86 | 0 | 0.4310 | 6.438 | 8.9 | 7.3967 | 7 | 330 | 19.1 | 377.07 | 3.59 | 24.8 |
| 0.08221 | 22.0 | 5.86 | 0 | 0.4310 | 6.957 | 6.8 | 8.9067 | 7 | 330 | 19.1 | 386.09 | 3.53 | 29.6 |
| 0.36894 | 22.0 | 5.86 | 0 | 0.4310 | 8.259 | 8.4 | 8.9067 | 7 | 330 | 19.1 | 396.90 | 3.54 | 42.8 |
| 0.04819 | 80.0 | 3.64 | 0 | 0.3920 | 6.108 | 32.0 | 9.2203 | 1 | 315 | 16.4 | 392.89 | 6.57 | 21.9 |
| 0.03548 | 80.0 | 3.64 | 0 | 0.3920 | 5.876 | 19.1 | 9.2203 | 1 | 315 | 16.4 | 395.18 | 9.25 | 20.9 |
| 0.01538 | 90.0 | 3.75 | 0 | 0.3940 | 7.454 | 34.2 | 6.3361 | 3 | 244 | 15.9 | 386.34 | 3.11 | 44.0 |
| 0.61154 | 20.0 | 3.97 | 0 | 0.6470 | 8.704 | 86.9 | 1.8010 | 5 | 264 | 13.0 | 389.70 | 5.12 | 50.0 |
| 0.66351 | 20.0 | 3.97 | 0 | 0.6470 | 7.333 | 100.0 | 1.8946 | 5 | 264 | 13.0 | 383.29 | 7.79 | 36.0 |
| 0.65665 | 20.0 | 3.97 | 0 | 0.6470 | 6.842 | 100.0 | 2.0107 | 5 | 264 | 13.0 | 391.93 | 6.90 | 30.1 |
| 0.54011 | 20.0 | 3.97 | 0 | 0.6470 | 7.203 | 81.8 | 2.1121 | 5 | 264 | 13.0 | 392.80 | 9.59 | 33.8 |
| 0.53412 | 20.0 | 3.97 | 0 | 0.6470 | 7.520 | 89.4 | 2.1398 | 5 | 264 | 13.0 | 388.37 | 7.26 | 43.1 |
| 0.52014 | 20.0 | 3.97 | 0 | 0.6470 | 8.398 | 91.5 | 2.2885 | 5 | 264 | 13.0 | 386.86 | 5.91 | 48.8 |
| 0.82526 | 20.0 | 3.97 | 0 | 0.6470 | 7.327 | 94.5 | 2.0788 | 5 | 264 | 13.0 | 393.42 | 11.25 | 31.0 |
| 0.55007 | 20.0 | 3.97 | 0 | 0.6470 | 7.206 | 91.6 | 1.9301 | 5 | 264 | 13.0 | 387.89 | 8.10 | 36.5 |
| 0.76162 | 20.0 | 3.97 | 0 | 0.6470 | 5.560 | 62.8 | 1.9865 | 5 | 264 | 13.0 | 392.40 | 10.45 | 22.8 |
| 0.78570 | 20.0 | 3.97 | 0 | 0.6470 | 7.014 | 84.6 | 2.1329 | 5 | 264 | 13.0 | 384.07 | 14.79 | 30.7 |
| 0.57834 | 20.0 | 3.97 | 0 | 0.5750 | 8.297 | 67.0 | 2.4216 | 5 | 264 | 13.0 | 384.54 | 7.44 | 50.0 |
| 0.54050 | 20.0 | 3.97 | 0 | 0.5750 | 7.470 | 52.6 | 2.8720 | 5 | 264 | 13.0 | 390.30 | 3.16 | 43.5 |
| 0.09065 | 20.0 | 6.96 | 1 | 0.4640 | 5.920 | 61.5 | 3.9175 | 3 | 223 | 18.6 | 391.34 | 13.65 | 20.7 |
| 0.29916 | 20.0 | 6.96 | 0 | 0.4640 | 5.856 | 42.1 | 4.4290 | 3 | 223 | 18.6 | 388.65 | 13.00 | 21.1 |
| 0.16211 | 20.0 | 6.96 | 0 | 0.4640 | 6.240 | 16.3 | 4.4290 | 3 | 223 | 18.6 | 396.90 | 6.59 | 25.2 |
| 0.11460 | 20.0 | 6.96 | 0 | 0.4640 | 6.538 | 58.7 | 3.9175 | 3 | 223 | 18.6 | 394.96 | 7.73 | 24.4 |
| 0.22188 | 20.0 | 6.96 | 1 | 0.4640 | 7.691 | 51.8 | 4.3665 | 3 | 223 | 18.6 | 390.77 | 6.58 | 35.2 |
| 0.05644 | 40.0 | 6.41 | 1 | 0.4470 | 6.758 | 32.9 | 4.0776 | 4 | 254 | 17.6 | 396.90 | 3.53 | 32.4 |
| 0.09604 | 40.0 | 6.41 | 0 | 0.4470 | 6.854 | 42.8 | 4.2673 | 4 | 254 | 17.6 | 396.90 | 2.98 | 32.0 |
| 0.10469 | 40.0 | 6.41 | 1 | 0.4470 | 7.267 | 49.0 | 4.7872 | 4 | 254 | 17.6 | 389.25 | 6.05 | 33.2 |
| 0.06127 | 40.0 | 6.41 | 1 | 0.4470 | 6.826 | 27.6 | 4.8628 | 4 | 254 | 17.6 | 393.45 | 4.16 | 33.1 |
| 0.07978 | 40.0 | 6.41 | 0 | 0.4470 | 6.482 | 32.1 | 4.1403 | 4 | 254 | 17.6 | 396.90 | 7.19 | 29.1 |
| 0.21038 | 20.0 | 3.33 | 0 | 0.4429 | 6.812 | 32.2 | 4.1007 | 5 | 216 | 14.9 | 396.90 | 4.85 | 35.1 |
| 0.03578 | 20.0 | 3.33 | 0 | 0.4429 | 7.820 | 64.5 | 4.6947 | 5 | 216 | 14.9 | 387.31 | 3.76 | 45.4 |
| 0.03705 | 20.0 | 3.33 | 0 | 0.4429 | 6.968 | 37.2 | 5.2447 | 5 | 216 | 14.9 | 392.23 | 4.59 | 35.4 |
| 0.06129 | 20.0 | 3.33 | 1 | 0.4429 | 7.645 | 49.7 | 5.2119 | 5 | 216 | 14.9 | 377.07 | 3.01 | 46.0 |
| 0.01501 | 90.0 | 1.21 | 1 | 0.4010 | 7.923 | 24.8 | 5.8850 | 1 | 198 | 13.6 | 395.52 | 3.16 | 50.0 |
| 0.00906 | 90.0 | 2.97 | 0 | 0.4000 | 7.088 | 20.8 | 7.3073 | 1 | 285 | 15.3 | 394.72 | 7.85 | 32.2 |
| 0.01096 | 55.0 | 2.25 | 0 | 0.3890 | 6.453 | 31.9 | 7.3073 | 1 | 300 | 15.3 | 394.72 | 8.23 | 22.0 |
| 0.01965 | 80.0 | 1.76 | 0 | 0.3850 | 6.230 | 31.5 | 9.0892 | 1 | 241 | 18.2 | 341.60 | 12.93 | 20.1 |
| 0.03871 | 52.5 | 5.32 | 0 | 0.4050 | 6.209 | 31.3 | 7.3172 | 6 | 293 | 16.6 | 396.90 | 7.14 | 23.2 |
| 0.04590 | 52.5 | 5.32 | 0 | 0.4050 | 6.315 | 45.6 | 7.3172 | 6 | 293 | 16.6 | 396.90 | 7.60 | 22.3 |
| 0.04297 | 52.5 | 5.32 | 0 | 0.4050 | 6.565 | 22.9 | 7.3172 | 6 | 293 | 16.6 | 371.72 | 9.51 | 24.8 |
| 0.03502 | 80.0 | 4.95 | 0 | 0.4110 | 6.861 | 27.9 | 5.1167 | 4 | 245 | 19.2 | 396.90 | 3.33 | 28.5 |
| 0.07886 | 80.0 | 4.95 | 0 | 0.4110 | 7.148 | 27.7 | 5.1167 | 4 | 245 | 19.2 | 396.90 | 3.56 | 37.3 |
| 0.03615 | 80.0 | 4.95 | 0 | 0.4110 | 6.630 | 23.4 | 5.1167 | 4 | 245 | 19.2 | 396.90 | 4.70 | 27.9 |
| 0.08265 | 0.0 | 13.92 | 0 | 0.4370 | 6.127 | 18.4 | 5.5027 | 4 | 289 | 16.0 | 396.90 | 8.58 | 23.9 |
| 0.08199 | 0.0 | 13.92 | 0 | 0.4370 | 6.009 | 42.3 | 5.5027 | 4 | 289 | 16.0 | 396.90 | 10.40 | 21.7 |
| 0.12932 | 0.0 | 13.92 | 0 | 0.4370 | 6.678 | 31.1 | 5.9604 | 4 | 289 | 16.0 | 396.90 | 6.27 | 28.6 |
| 0.05372 | 0.0 | 13.92 | 0 | 0.4370 | 6.549 | 51.0 | 5.9604 | 4 | 289 | 16.0 | 392.85 | 7.39 | 27.1 |
| 0.14103 | 0.0 | 13.92 | 0 | 0.4370 | 5.790 | 58.0 | 6.3200 | 4 | 289 | 16.0 | 396.90 | 15.84 | 20.3 |
| 0.06466 | 70.0 | 2.24 | 0 | 0.4000 | 6.345 | 20.1 | 7.8278 | 5 | 358 | 14.8 | 368.24 | 4.97 | 22.5 |
| 0.05561 | 70.0 | 2.24 | 0 | 0.4000 | 7.041 | 10.0 | 7.8278 | 5 | 358 | 14.8 | 371.58 | 4.74 | 29.0 |
| 0.04417 | 70.0 | 2.24 | 0 | 0.4000 | 6.871 | 47.4 | 7.8278 | 5 | 358 | 14.8 | 390.86 | 6.07 | 24.8 |
| 0.03537 | 34.0 | 6.09 | 0 | 0.4330 | 6.590 | 40.4 | 5.4917 | 7 | 329 | 16.1 | 395.75 | 9.50 | 22.0 |
| 0.09266 | 34.0 | 6.09 | 0 | 0.4330 | 6.495 | 18.4 | 5.4917 | 7 | 329 | 16.1 | 383.61 | 8.67 | 26.4 |
| 0.10000 | 34.0 | 6.09 | 0 | 0.4330 | 6.982 | 17.7 | 5.4917 | 7 | 329 | 16.1 | 390.43 | 4.86 | 33.1 |
| 0.05515 | 33.0 | 2.18 | 0 | 0.4720 | 7.236 | 41.1 | 4.0220 | 7 | 222 | 18.4 | 393.68 | 6.93 | 36.1 |
| 0.05479 | 33.0 | 2.18 | 0 | 0.4720 | 6.616 | 58.1 | 3.3700 | 7 | 222 | 18.4 | 393.36 | 8.93 | 28.4 |
| 0.07503 | 33.0 | 2.18 | 0 | 0.4720 | 7.420 | 71.9 | 3.0992 | 7 | 222 | 18.4 | 396.90 | 6.47 | 33.4 |
| 0.04932 | 33.0 | 2.18 | 0 | 0.4720 | 6.849 | 70.3 | 3.1827 | 7 | 222 | 18.4 | 396.90 | 7.53 | 28.2 |
| 0.49298 | 0.0 | 9.90 | 0 | 0.5440 | 6.635 | 82.5 | 3.3175 | 4 | 304 | 18.4 | 396.90 | 4.54 | 22.8 |
| 0.34940 | 0.0 | 9.90 | 0 | 0.5440 | 5.972 | 76.7 | 3.1025 | 4 | 304 | 18.4 | 396.24 | 9.97 | 20.3 |
| 2.63548 | 0.0 | 9.90 | 0 | 0.5440 | 4.973 | 37.8 | 2.5194 | 4 | 304 | 18.4 | 350.45 | 12.64 | 16.1 |
| 0.79041 | 0.0 | 9.90 | 0 | 0.5440 | 6.122 | 52.8 | 2.6403 | 4 | 304 | 18.4 | 396.90 | 5.98 | 22.1 |
| 0.26169 | 0.0 | 9.90 | 0 | 0.5440 | 6.023 | 90.4 | 2.8340 | 4 | 304 | 18.4 | 396.30 | 11.72 | 19.4 |
| 0.26938 | 0.0 | 9.90 | 0 | 0.5440 | 6.266 | 82.8 | 3.2628 | 4 | 304 | 18.4 | 393.39 | 7.90 | 21.6 |
| 0.36920 | 0.0 | 9.90 | 0 | 0.5440 | 6.567 | 87.3 | 3.6023 | 4 | 304 | 18.4 | 395.69 | 9.28 | 23.8 |
| 0.25356 | 0.0 | 9.90 | 0 | 0.5440 | 5.705 | 77.7 | 3.9450 | 4 | 304 | 18.4 | 396.42 | 11.50 | 16.2 |
| 0.31827 | 0.0 | 9.90 | 0 | 0.5440 | 5.914 | 83.2 | 3.9986 | 4 | 304 | 18.4 | 390.70 | 18.33 | 17.8 |
| 0.24522 | 0.0 | 9.90 | 0 | 0.5440 | 5.782 | 71.7 | 4.0317 | 4 | 304 | 18.4 | 396.90 | 15.94 | 19.8 |
| 0.40202 | 0.0 | 9.90 | 0 | 0.5440 | 6.382 | 67.2 | 3.5325 | 4 | 304 | 18.4 | 395.21 | 10.36 | 23.1 |
| 0.47547 | 0.0 | 9.90 | 0 | 0.5440 | 6.113 | 58.8 | 4.0019 | 4 | 304 | 18.4 | 396.23 | 12.73 | 21.0 |
| 0.16760 | 0.0 | 7.38 | 0 | 0.4930 | 6.426 | 52.3 | 4.5404 | 5 | 287 | 19.6 | 396.90 | 7.20 | 23.8 |
| 0.18159 | 0.0 | 7.38 | 0 | 0.4930 | 6.376 | 54.3 | 4.5404 | 5 | 287 | 19.6 | 396.90 | 6.87 | 23.1 |
| 0.35114 | 0.0 | 7.38 | 0 | 0.4930 | 6.041 | 49.9 | 4.7211 | 5 | 287 | 19.6 | 396.90 | 7.70 | 20.4 |
| 0.28392 | 0.0 | 7.38 | 0 | 0.4930 | 5.708 | 74.3 | 4.7211 | 5 | 287 | 19.6 | 391.13 | 11.74 | 18.5 |
| 0.34109 | 0.0 | 7.38 | 0 | 0.4930 | 6.415 | 40.1 | 4.7211 | 5 | 287 | 19.6 | 396.90 | 6.12 | 25.0 |
| 0.19186 | 0.0 | 7.38 | 0 | 0.4930 | 6.431 | 14.7 | 5.4159 | 5 | 287 | 19.6 | 393.68 | 5.08 | 24.6 |
| 0.30347 | 0.0 | 7.38 | 0 | 0.4930 | 6.312 | 28.9 | 5.4159 | 5 | 287 | 19.6 | 396.90 | 6.15 | 23.0 |
| 0.24103 | 0.0 | 7.38 | 0 | 0.4930 | 6.083 | 43.7 | 5.4159 | 5 | 287 | 19.6 | 396.90 | 12.79 | 22.2 |
| 0.06617 | 0.0 | 3.24 | 0 | 0.4600 | 5.868 | 25.8 | 5.2146 | 4 | 430 | 16.9 | 382.44 | 9.97 | 19.3 |
| 0.06724 | 0.0 | 3.24 | 0 | 0.4600 | 6.333 | 17.2 | 5.2146 | 4 | 430 | 16.9 | 375.21 | 7.34 | 22.6 |
| 0.04544 | 0.0 | 3.24 | 0 | 0.4600 | 6.144 | 32.2 | 5.8736 | 4 | 430 | 16.9 | 368.57 | 9.09 | 19.8 |
| 0.05023 | 35.0 | 6.06 | 0 | 0.4379 | 5.706 | 28.4 | 6.6407 | 1 | 304 | 16.9 | 394.02 | 12.43 | 17.1 |
| 0.03466 | 35.0 | 6.06 | 0 | 0.4379 | 6.031 | 23.3 | 6.6407 | 1 | 304 | 16.9 | 362.25 | 7.83 | 19.4 |
| 0.05083 | 0.0 | 5.19 | 0 | 0.5150 | 6.316 | 38.1 | 6.4584 | 5 | 224 | 20.2 | 389.71 | 5.68 | 22.2 |
| 0.03738 | 0.0 | 5.19 | 0 | 0.5150 | 6.310 | 38.5 | 6.4584 | 5 | 224 | 20.2 | 389.40 | 6.75 | 20.7 |
| 0.03961 | 0.0 | 5.19 | 0 | 0.5150 | 6.037 | 34.5 | 5.9853 | 5 | 224 | 20.2 | 396.90 | 8.01 | 21.1 |
| 0.03427 | 0.0 | 5.19 | 0 | 0.5150 | 5.869 | 46.3 | 5.2311 | 5 | 224 | 20.2 | 396.90 | 9.80 | 19.5 |
| 0.03041 | 0.0 | 5.19 | 0 | 0.5150 | 5.895 | 59.6 | 5.6150 | 5 | 224 | 20.2 | 394.81 | 10.56 | 18.5 |
| 0.03306 | 0.0 | 5.19 | 0 | 0.5150 | 6.059 | 37.3 | 4.8122 | 5 | 224 | 20.2 | 396.14 | 8.51 | 20.6 |
| 0.05497 | 0.0 | 5.19 | 0 | 0.5150 | 5.985 | 45.4 | 4.8122 | 5 | 224 | 20.2 | 396.90 | 9.74 | 19.0 |
| 0.06151 | 0.0 | 5.19 | 0 | 0.5150 | 5.968 | 58.5 | 4.8122 | 5 | 224 | 20.2 | 396.90 | 9.29 | 18.7 |
| 0.01301 | 35.0 | 1.52 | 0 | 0.4420 | 7.241 | 49.3 | 7.0379 | 1 | 284 | 15.5 | 394.74 | 5.49 | 32.7 |
| 0.02498 | 0.0 | 1.89 | 0 | 0.5180 | 6.540 | 59.7 | 6.2669 | 1 | 422 | 15.9 | 389.96 | 8.65 | 16.5 |
| 0.02543 | 55.0 | 3.78 | 0 | 0.4840 | 6.696 | 56.4 | 5.7321 | 5 | 370 | 17.6 | 396.90 | 7.18 | 23.9 |
| 0.03049 | 55.0 | 3.78 | 0 | 0.4840 | 6.874 | 28.1 | 6.4654 | 5 | 370 | 17.6 | 387.97 | 4.61 | 31.2 |
| 0.03113 | 0.0 | 4.39 | 0 | 0.4420 | 6.014 | 48.5 | 8.0136 | 3 | 352 | 18.8 | 385.64 | 10.53 | 17.5 |
| 0.06162 | 0.0 | 4.39 | 0 | 0.4420 | 5.898 | 52.3 | 8.0136 | 3 | 352 | 18.8 | 364.61 | 12.67 | 17.2 |
| 0.01870 | 85.0 | 4.15 | 0 | 0.4290 | 6.516 | 27.7 | 8.5353 | 4 | 351 | 17.9 | 392.43 | 6.36 | 23.1 |
| 0.01501 | 80.0 | 2.01 | 0 | 0.4350 | 6.635 | 29.7 | 8.3440 | 4 | 280 | 17.0 | 390.94 | 5.99 | 24.5 |
| 0.02899 | 40.0 | 1.25 | 0 | 0.4290 | 6.939 | 34.5 | 8.7921 | 1 | 335 | 19.7 | 389.85 | 5.89 | 26.6 |
| 0.06211 | 40.0 | 1.25 | 0 | 0.4290 | 6.490 | 44.4 | 8.7921 | 1 | 335 | 19.7 | 396.90 | 5.98 | 22.9 |
| 0.07950 | 60.0 | 1.69 | 0 | 0.4110 | 6.579 | 35.9 | 10.7103 | 4 | 411 | 18.3 | 370.78 | 5.49 | 24.1 |
| 0.07244 | 60.0 | 1.69 | 0 | 0.4110 | 5.884 | 18.5 | 10.7103 | 4 | 411 | 18.3 | 392.33 | 7.79 | 18.6 |
| 0.01709 | 90.0 | 2.02 | 0 | 0.4100 | 6.728 | 36.1 | 12.1265 | 5 | 187 | 17.0 | 384.46 | 4.50 | 30.1 |
| 0.04301 | 80.0 | 1.91 | 0 | 0.4130 | 5.663 | 21.9 | 10.5857 | 4 | 334 | 22.0 | 382.80 | 8.05 | 18.2 |
| 0.10659 | 80.0 | 1.91 | 0 | 0.4130 | 5.936 | 19.5 | 10.5857 | 4 | 334 | 22.0 | 376.04 | 5.57 | 20.6 |
| 8.98296 | 0.0 | 18.10 | 1 | 0.7700 | 6.212 | 97.4 | 2.1222 | 24 | 666 | 20.2 | 377.73 | 17.60 | 17.8 |
| 3.84970 | 0.0 | 18.10 | 1 | 0.7700 | 6.395 | 91.0 | 2.5052 | 24 | 666 | 20.2 | 391.34 | 13.27 | 21.7 |
| 5.20177 | 0.0 | 18.10 | 1 | 0.7700 | 6.127 | 83.4 | 2.7227 | 24 | 666 | 20.2 | 395.43 | 11.48 | 22.7 |
| 4.26131 | 0.0 | 18.10 | 0 | 0.7700 | 6.112 | 81.3 | 2.5091 | 24 | 666 | 20.2 | 390.74 | 12.67 | 22.6 |
| 4.54192 | 0.0 | 18.10 | 0 | 0.7700 | 6.398 | 88.0 | 2.5182 | 24 | 666 | 20.2 | 374.56 | 7.79 | 25.0 |
| 3.83684 | 0.0 | 18.10 | 0 | 0.7700 | 6.251 | 91.1 | 2.2955 | 24 | 666 | 20.2 | 350.65 | 14.19 | 19.9 |
| 3.67822 | 0.0 | 18.10 | 0 | 0.7700 | 5.362 | 96.2 | 2.1036 | 24 | 666 | 20.2 | 380.79 | 10.19 | 20.8 |
| 4.22239 | 0.0 | 18.10 | 1 | 0.7700 | 5.803 | 89.0 | 1.9047 | 24 | 666 | 20.2 | 353.04 | 14.64 | 16.8 |
| 3.47428 | 0.0 | 18.10 | 1 | 0.7180 | 8.780 | 82.9 | 1.9047 | 24 | 666 | 20.2 | 354.55 | 5.29 | 21.9 |
| 4.55587 | 0.0 | 18.10 | 0 | 0.7180 | 3.561 | 87.9 | 1.6132 | 24 | 666 | 20.2 | 354.70 | 7.12 | 27.5 |
| 3.69695 | 0.0 | 18.10 | 0 | 0.7180 | 4.963 | 91.4 | 1.7523 | 24 | 666 | 20.2 | 316.03 | 14.00 | 21.9 |
| 13.52220 | 0.0 | 18.10 | 0 | 0.6310 | 3.863 | 100.0 | 1.5106 | 24 | 666 | 20.2 | 131.42 | 13.33 | 23.1 |
| 4.89822 | 0.0 | 18.10 | 0 | 0.6310 | 4.970 | 100.0 | 1.3325 | 24 | 666 | 20.2 | 375.52 | 3.26 | 50.0 |
| 5.66998 | 0.0 | 18.10 | 1 | 0.6310 | 6.683 | 96.8 | 1.3567 | 24 | 666 | 20.2 | 375.33 | 3.73 | 50.0 |
| 6.53876 | 0.0 | 18.10 | 1 | 0.6310 | 7.016 | 97.5 | 1.2024 | 24 | 666 | 20.2 | 392.05 | 2.96 | 50.0 |
| 9.23230 | 0.0 | 18.10 | 0 | 0.6310 | 6.216 | 100.0 | 1.1691 | 24 | 666 | 20.2 | 366.15 | 9.53 | 50.0 |
| 8.26725 | 0.0 | 18.10 | 1 | 0.6680 | 5.875 | 89.6 | 1.1296 | 24 | 666 | 20.2 | 347.88 | 8.88 | 50.0 |
| 11.10810 | 0.0 | 18.10 | 0 | 0.6680 | 4.906 | 100.0 | 1.1742 | 24 | 666 | 20.2 | 396.90 | 34.77 | 13.8 |
| 18.49820 | 0.0 | 18.10 | 0 | 0.6680 | 4.138 | 100.0 | 1.1370 | 24 | 666 | 20.2 | 396.90 | 37.97 | 13.8 |
| 19.60910 | 0.0 | 18.10 | 0 | 0.6710 | 7.313 | 97.9 | 1.3163 | 24 | 666 | 20.2 | 396.90 | 13.44 | 15.0 |
| 15.28800 | 0.0 | 18.10 | 0 | 0.6710 | 6.649 | 93.3 | 1.3449 | 24 | 666 | 20.2 | 363.02 | 23.24 | 13.9 |
| 9.82349 | 0.0 | 18.10 | 0 | 0.6710 | 6.794 | 98.8 | 1.3580 | 24 | 666 | 20.2 | 396.90 | 21.24 | 13.3 |
| 23.64820 | 0.0 | 18.10 | 0 | 0.6710 | 6.380 | 96.2 | 1.3861 | 24 | 666 | 20.2 | 396.90 | 23.69 | 13.1 |
| 17.86670 | 0.0 | 18.10 | 0 | 0.6710 | 6.223 | 100.0 | 1.3861 | 24 | 666 | 20.2 | 393.74 | 21.78 | 10.2 |
| 88.97620 | 0.0 | 18.10 | 0 | 0.6710 | 6.968 | 91.9 | 1.4165 | 24 | 666 | 20.2 | 396.90 | 17.21 | 10.4 |
| 15.87440 | 0.0 | 18.10 | 0 | 0.6710 | 6.545 | 99.1 | 1.5192 | 24 | 666 | 20.2 | 396.90 | 21.08 | 10.9 |
| 9.18702 | 0.0 | 18.10 | 0 | 0.7000 | 5.536 | 100.0 | 1.5804 | 24 | 666 | 20.2 | 396.90 | 23.60 | 11.3 |
| 7.99248 | 0.0 | 18.10 | 0 | 0.7000 | 5.520 | 100.0 | 1.5331 | 24 | 666 | 20.2 | 396.90 | 24.56 | 12.3 |
| 20.08490 | 0.0 | 18.10 | 0 | 0.7000 | 4.368 | 91.2 | 1.4395 | 24 | 666 | 20.2 | 285.83 | 30.63 | 8.8 |
| 16.81180 | 0.0 | 18.10 | 0 | 0.7000 | 5.277 | 98.1 | 1.4261 | 24 | 666 | 20.2 | 396.90 | 30.81 | 7.2 |
| 24.39380 | 0.0 | 18.10 | 0 | 0.7000 | 4.652 | 100.0 | 1.4672 | 24 | 666 | 20.2 | 396.90 | 28.28 | 10.5 |
| 22.59710 | 0.0 | 18.10 | 0 | 0.7000 | 5.000 | 89.5 | 1.5184 | 24 | 666 | 20.2 | 396.90 | 31.99 | 7.4 |
| 14.33370 | 0.0 | 18.10 | 0 | 0.7000 | 4.880 | 100.0 | 1.5895 | 24 | 666 | 20.2 | 372.92 | 30.62 | 10.2 |
| 8.15174 | 0.0 | 18.10 | 0 | 0.7000 | 5.390 | 98.9 | 1.7281 | 24 | 666 | 20.2 | 396.90 | 20.85 | 11.5 |
| 6.96215 | 0.0 | 18.10 | 0 | 0.7000 | 5.713 | 97.0 | 1.9265 | 24 | 666 | 20.2 | 394.43 | 17.11 | 15.1 |
| 5.29305 | 0.0 | 18.10 | 0 | 0.7000 | 6.051 | 82.5 | 2.1678 | 24 | 666 | 20.2 | 378.38 | 18.76 | 23.2 |
| 11.57790 | 0.0 | 18.10 | 0 | 0.7000 | 5.036 | 97.0 | 1.7700 | 24 | 666 | 20.2 | 396.90 | 25.68 | 9.7 |
| 8.64476 | 0.0 | 18.10 | 0 | 0.6930 | 6.193 | 92.6 | 1.7912 | 24 | 666 | 20.2 | 396.90 | 15.17 | 13.8 |
| 13.35980 | 0.0 | 18.10 | 0 | 0.6930 | 5.887 | 94.7 | 1.7821 | 24 | 666 | 20.2 | 396.90 | 16.35 | 12.7 |
| 8.71675 | 0.0 | 18.10 | 0 | 0.6930 | 6.471 | 98.8 | 1.7257 | 24 | 666 | 20.2 | 391.98 | 17.12 | 13.1 |
| 5.87205 | 0.0 | 18.10 | 0 | 0.6930 | 6.405 | 96.0 | 1.6768 | 24 | 666 | 20.2 | 396.90 | 19.37 | 12.5 |
| 7.67202 | 0.0 | 18.10 | 0 | 0.6930 | 5.747 | 98.9 | 1.6334 | 24 | 666 | 20.2 | 393.10 | 19.92 | 8.5 |
| 38.35180 | 0.0 | 18.10 | 0 | 0.6930 | 5.453 | 100.0 | 1.4896 | 24 | 666 | 20.2 | 396.90 | 30.59 | 5.0 |
| 9.91655 | 0.0 | 18.10 | 0 | 0.6930 | 5.852 | 77.8 | 1.5004 | 24 | 666 | 20.2 | 338.16 | 29.97 | 6.3 |
| 25.04610 | 0.0 | 18.10 | 0 | 0.6930 | 5.987 | 100.0 | 1.5888 | 24 | 666 | 20.2 | 396.90 | 26.77 | 5.6 |
| 14.23620 | 0.0 | 18.10 | 0 | 0.6930 | 6.343 | 100.0 | 1.5741 | 24 | 666 | 20.2 | 396.90 | 20.32 | 7.2 |
| 9.59571 | 0.0 | 18.10 | 0 | 0.6930 | 6.404 | 100.0 | 1.6390 | 24 | 666 | 20.2 | 376.11 | 20.31 | 12.1 |
| 24.80170 | 0.0 | 18.10 | 0 | 0.6930 | 5.349 | 96.0 | 1.7028 | 24 | 666 | 20.2 | 396.90 | 19.77 | 8.3 |
| 41.52920 | 0.0 | 18.10 | 0 | 0.6930 | 5.531 | 85.4 | 1.6074 | 24 | 666 | 20.2 | 329.46 | 27.38 | 8.5 |
| 67.92080 | 0.0 | 18.10 | 0 | 0.6930 | 5.683 | 100.0 | 1.4254 | 24 | 666 | 20.2 | 384.97 | 22.98 | 5.0 |
| 20.71620 | 0.0 | 18.10 | 0 | 0.6590 | 4.138 | 100.0 | 1.1781 | 24 | 666 | 20.2 | 370.22 | 23.34 | 11.9 |
| 11.95110 | 0.0 | 18.10 | 0 | 0.6590 | 5.608 | 100.0 | 1.2852 | 24 | 666 | 20.2 | 332.09 | 12.13 | 27.9 |
| 7.40389 | 0.0 | 18.10 | 0 | 0.5970 | 5.617 | 97.9 | 1.4547 | 24 | 666 | 20.2 | 314.64 | 26.40 | 17.2 |
| 14.43830 | 0.0 | 18.10 | 0 | 0.5970 | 6.852 | 100.0 | 1.4655 | 24 | 666 | 20.2 | 179.36 | 19.78 | 27.5 |
| 51.13580 | 0.0 | 18.10 | 0 | 0.5970 | 5.757 | 100.0 | 1.4130 | 24 | 666 | 20.2 | 2.60 | 10.11 | 15.0 |
| 14.05070 | 0.0 | 18.10 | 0 | 0.5970 | 6.657 | 100.0 | 1.5275 | 24 | 666 | 20.2 | 35.05 | 21.22 | 17.2 |
| 18.81100 | 0.0 | 18.10 | 0 | 0.5970 | 4.628 | 100.0 | 1.5539 | 24 | 666 | 20.2 | 28.79 | 34.37 | 17.9 |
| 28.65580 | 0.0 | 18.10 | 0 | 0.5970 | 5.155 | 100.0 | 1.5894 | 24 | 666 | 20.2 | 210.97 | 20.08 | 16.3 |
| 45.74610 | 0.0 | 18.10 | 0 | 0.6930 | 4.519 | 100.0 | 1.6582 | 24 | 666 | 20.2 | 88.27 | 36.98 | 7.0 |
| 18.08460 | 0.0 | 18.10 | 0 | 0.6790 | 6.434 | 100.0 | 1.8347 | 24 | 666 | 20.2 | 27.25 | 29.05 | 7.2 |
| 10.83420 | 0.0 | 18.10 | 0 | 0.6790 | 6.782 | 90.8 | 1.8195 | 24 | 666 | 20.2 | 21.57 | 25.79 | 7.5 |
| 25.94060 | 0.0 | 18.10 | 0 | 0.6790 | 5.304 | 89.1 | 1.6475 | 24 | 666 | 20.2 | 127.36 | 26.64 | 10.4 |
| 73.53410 | 0.0 | 18.10 | 0 | 0.6790 | 5.957 | 100.0 | 1.8026 | 24 | 666 | 20.2 | 16.45 | 20.62 | 8.8 |
| 11.81230 | 0.0 | 18.10 | 0 | 0.7180 | 6.824 | 76.5 | 1.7940 | 24 | 666 | 20.2 | 48.45 | 22.74 | 8.4 |
| 11.08740 | 0.0 | 18.10 | 0 | 0.7180 | 6.411 | 100.0 | 1.8589 | 24 | 666 | 20.2 | 318.75 | 15.02 | 16.7 |
| 7.02259 | 0.0 | 18.10 | 0 | 0.7180 | 6.006 | 95.3 | 1.8746 | 24 | 666 | 20.2 | 319.98 | 15.70 | 14.2 |
| 12.04820 | 0.0 | 18.10 | 0 | 0.6140 | 5.648 | 87.6 | 1.9512 | 24 | 666 | 20.2 | 291.55 | 14.10 | 20.8 |
| 7.05042 | 0.0 | 18.10 | 0 | 0.6140 | 6.103 | 85.1 | 2.0218 | 24 | 666 | 20.2 | 2.52 | 23.29 | 13.4 |
| 8.79212 | 0.0 | 18.10 | 0 | 0.5840 | 5.565 | 70.6 | 2.0635 | 24 | 666 | 20.2 | 3.65 | 17.16 | 11.7 |
| 15.86030 | 0.0 | 18.10 | 0 | 0.6790 | 5.896 | 95.4 | 1.9096 | 24 | 666 | 20.2 | 7.68 | 24.39 | 8.3 |
| 12.24720 | 0.0 | 18.10 | 0 | 0.5840 | 5.837 | 59.7 | 1.9976 | 24 | 666 | 20.2 | 24.65 | 15.69 | 10.2 |
| 37.66190 | 0.0 | 18.10 | 0 | 0.6790 | 6.202 | 78.7 | 1.8629 | 24 | 666 | 20.2 | 18.82 | 14.52 | 10.9 |
| 7.36711 | 0.0 | 18.10 | 0 | 0.6790 | 6.193 | 78.1 | 1.9356 | 24 | 666 | 20.2 | 96.73 | 21.52 | 11.0 |
| 9.33889 | 0.0 | 18.10 | 0 | 0.6790 | 6.380 | 95.6 | 1.9682 | 24 | 666 | 20.2 | 60.72 | 24.08 | 9.5 |
| 8.49213 | 0.0 | 18.10 | 0 | 0.5840 | 6.348 | 86.1 | 2.0527 | 24 | 666 | 20.2 | 83.45 | 17.64 | 14.5 |
| 10.06230 | 0.0 | 18.10 | 0 | 0.5840 | 6.833 | 94.3 | 2.0882 | 24 | 666 | 20.2 | 81.33 | 19.69 | 14.1 |
| 6.44405 | 0.0 | 18.10 | 0 | 0.5840 | 6.425 | 74.8 | 2.2004 | 24 | 666 | 20.2 | 97.95 | 12.03 | 16.1 |
| 5.58107 | 0.0 | 18.10 | 0 | 0.7130 | 6.436 | 87.9 | 2.3158 | 24 | 666 | 20.2 | 100.19 | 16.22 | 14.3 |
| 13.91340 | 0.0 | 18.10 | 0 | 0.7130 | 6.208 | 95.0 | 2.2222 | 24 | 666 | 20.2 | 100.63 | 15.17 | 11.7 |
| 11.16040 | 0.0 | 18.10 | 0 | 0.7400 | 6.629 | 94.6 | 2.1247 | 24 | 666 | 20.2 | 109.85 | 23.27 | 13.4 |
| 14.42080 | 0.0 | 18.10 | 0 | 0.7400 | 6.461 | 93.3 | 2.0026 | 24 | 666 | 20.2 | 27.49 | 18.05 | 9.6 |
| 15.17720 | 0.0 | 18.10 | 0 | 0.7400 | 6.152 | 100.0 | 1.9142 | 24 | 666 | 20.2 | 9.32 | 26.45 | 8.7 |
| 13.67810 | 0.0 | 18.10 | 0 | 0.7400 | 5.935 | 87.9 | 1.8206 | 24 | 666 | 20.2 | 68.95 | 34.02 | 8.4 |
| 9.39063 | 0.0 | 18.10 | 0 | 0.7400 | 5.627 | 93.9 | 1.8172 | 24 | 666 | 20.2 | 396.90 | 22.88 | 12.8 |
| 22.05110 | 0.0 | 18.10 | 0 | 0.7400 | 5.818 | 92.4 | 1.8662 | 24 | 666 | 20.2 | 391.45 | 22.11 | 10.5 |
| 9.72418 | 0.0 | 18.10 | 0 | 0.7400 | 6.406 | 97.2 | 2.0651 | 24 | 666 | 20.2 | 385.96 | 19.52 | 17.1 |
| 5.66637 | 0.0 | 18.10 | 0 | 0.7400 | 6.219 | 100.0 | 2.0048 | 24 | 666 | 20.2 | 395.69 | 16.59 | 18.4 |
| 9.96654 | 0.0 | 18.10 | 0 | 0.7400 | 6.485 | 100.0 | 1.9784 | 24 | 666 | 20.2 | 386.73 | 18.85 | 15.4 |
| 12.80230 | 0.0 | 18.10 | 0 | 0.7400 | 5.854 | 96.6 | 1.8956 | 24 | 666 | 20.2 | 240.52 | 23.79 | 10.8 |
| 10.67180 | 0.0 | 18.10 | 0 | 0.7400 | 6.459 | 94.8 | 1.9879 | 24 | 666 | 20.2 | 43.06 | 23.98 | 11.8 |
| 6.28807 | 0.0 | 18.10 | 0 | 0.7400 | 6.341 | 96.4 | 2.0720 | 24 | 666 | 20.2 | 318.01 | 17.79 | 14.9 |
| 9.92485 | 0.0 | 18.10 | 0 | 0.7400 | 6.251 | 96.6 | 2.1980 | 24 | 666 | 20.2 | 388.52 | 16.44 | 12.6 |
| 9.32909 | 0.0 | 18.10 | 0 | 0.7130 | 6.185 | 98.7 | 2.2616 | 24 | 666 | 20.2 | 396.90 | 18.13 | 14.1 |
| 7.52601 | 0.0 | 18.10 | 0 | 0.7130 | 6.417 | 98.3 | 2.1850 | 24 | 666 | 20.2 | 304.21 | 19.31 | 13.0 |
| 6.71772 | 0.0 | 18.10 | 0 | 0.7130 | 6.749 | 92.6 | 2.3236 | 24 | 666 | 20.2 | 0.32 | 17.44 | 13.4 |
| 5.44114 | 0.0 | 18.10 | 0 | 0.7130 | 6.655 | 98.2 | 2.3552 | 24 | 666 | 20.2 | 355.29 | 17.73 | 15.2 |
| 5.09017 | 0.0 | 18.10 | 0 | 0.7130 | 6.297 | 91.8 | 2.3682 | 24 | 666 | 20.2 | 385.09 | 17.27 | 16.1 |
| 8.24809 | 0.0 | 18.10 | 0 | 0.7130 | 7.393 | 99.3 | 2.4527 | 24 | 666 | 20.2 | 375.87 | 16.74 | 17.8 |
| 9.51363 | 0.0 | 18.10 | 0 | 0.7130 | 6.728 | 94.1 | 2.4961 | 24 | 666 | 20.2 | 6.68 | 18.71 | 14.9 |
| 4.75237 | 0.0 | 18.10 | 0 | 0.7130 | 6.525 | 86.5 | 2.4358 | 24 | 666 | 20.2 | 50.92 | 18.13 | 14.1 |
| 4.66883 | 0.0 | 18.10 | 0 | 0.7130 | 5.976 | 87.9 | 2.5806 | 24 | 666 | 20.2 | 10.48 | 19.01 | 12.7 |
| 8.20058 | 0.0 | 18.10 | 0 | 0.7130 | 5.936 | 80.3 | 2.7792 | 24 | 666 | 20.2 | 3.50 | 16.94 | 13.5 |
| 7.75223 | 0.0 | 18.10 | 0 | 0.7130 | 6.301 | 83.7 | 2.7831 | 24 | 666 | 20.2 | 272.21 | 16.23 | 14.9 |
| 6.80117 | 0.0 | 18.10 | 0 | 0.7130 | 6.081 | 84.4 | 2.7175 | 24 | 666 | 20.2 | 396.90 | 14.70 | 20.0 |
| 4.81213 | 0.0 | 18.10 | 0 | 0.7130 | 6.701 | 90.0 | 2.5975 | 24 | 666 | 20.2 | 255.23 | 16.42 | 16.4 |
| 3.69311 | 0.0 | 18.10 | 0 | 0.7130 | 6.376 | 88.4 | 2.5671 | 24 | 666 | 20.2 | 391.43 | 14.65 | 17.7 |
| 6.65492 | 0.0 | 18.10 | 0 | 0.7130 | 6.317 | 83.0 | 2.7344 | 24 | 666 | 20.2 | 396.90 | 13.99 | 19.5 |
| 5.82115 | 0.0 | 18.10 | 0 | 0.7130 | 6.513 | 89.9 | 2.8016 | 24 | 666 | 20.2 | 393.82 | 10.29 | 20.2 |
| 7.83932 | 0.0 | 18.10 | 0 | 0.6550 | 6.209 | 65.4 | 2.9634 | 24 | 666 | 20.2 | 396.90 | 13.22 | 21.4 |
| 3.16360 | 0.0 | 18.10 | 0 | 0.6550 | 5.759 | 48.2 | 3.0665 | 24 | 666 | 20.2 | 334.40 | 14.13 | 19.9 |
| 3.77498 | 0.0 | 18.10 | 0 | 0.6550 | 5.952 | 84.7 | 2.8715 | 24 | 666 | 20.2 | 22.01 | 17.15 | 19.0 |
| 4.42228 | 0.0 | 18.10 | 0 | 0.5840 | 6.003 | 94.5 | 2.5403 | 24 | 666 | 20.2 | 331.29 | 21.32 | 19.1 |
| 15.57570 | 0.0 | 18.10 | 0 | 0.5800 | 5.926 | 71.0 | 2.9084 | 24 | 666 | 20.2 | 368.74 | 18.13 | 19.1 |
| 13.07510 | 0.0 | 18.10 | 0 | 0.5800 | 5.713 | 56.7 | 2.8237 | 24 | 666 | 20.2 | 396.90 | 14.76 | 20.1 |
| 4.34879 | 0.0 | 18.10 | 0 | 0.5800 | 6.167 | 84.0 | 3.0334 | 24 | 666 | 20.2 | 396.90 | 16.29 | 19.9 |
| 4.03841 | 0.0 | 18.10 | 0 | 0.5320 | 6.229 | 90.7 | 3.0993 | 24 | 666 | 20.2 | 395.33 | 12.87 | 19.6 |
| 3.56868 | 0.0 | 18.10 | 0 | 0.5800 | 6.437 | 75.0 | 2.8965 | 24 | 666 | 20.2 | 393.37 | 14.36 | 23.2 |
| 4.64689 | 0.0 | 18.10 | 0 | 0.6140 | 6.980 | 67.6 | 2.5329 | 24 | 666 | 20.2 | 374.68 | 11.66 | 29.8 |
| 8.05579 | 0.0 | 18.10 | 0 | 0.5840 | 5.427 | 95.4 | 2.4298 | 24 | 666 | 20.2 | 352.58 | 18.14 | 13.8 |
| 6.39312 | 0.0 | 18.10 | 0 | 0.5840 | 6.162 | 97.4 | 2.2060 | 24 | 666 | 20.2 | 302.76 | 24.10 | 13.3 |
| 4.87141 | 0.0 | 18.10 | 0 | 0.6140 | 6.484 | 93.6 | 2.3053 | 24 | 666 | 20.2 | 396.21 | 18.68 | 16.7 |
| 15.02340 | 0.0 | 18.10 | 0 | 0.6140 | 5.304 | 97.3 | 2.1007 | 24 | 666 | 20.2 | 349.48 | 24.91 | 12.0 |
| 10.23300 | 0.0 | 18.10 | 0 | 0.6140 | 6.185 | 96.7 | 2.1705 | 24 | 666 | 20.2 | 379.70 | 18.03 | 14.6 |
| 14.33370 | 0.0 | 18.10 | 0 | 0.6140 | 6.229 | 88.0 | 1.9512 | 24 | 666 | 20.2 | 383.32 | 13.11 | 21.4 |
| 5.82401 | 0.0 | 18.10 | 0 | 0.5320 | 6.242 | 64.7 | 3.4242 | 24 | 666 | 20.2 | 396.90 | 10.74 | 23.0 |
| 5.70818 | 0.0 | 18.10 | 0 | 0.5320 | 6.750 | 74.9 | 3.3317 | 24 | 666 | 20.2 | 393.07 | 7.74 | 23.7 |
| 5.73116 | 0.0 | 18.10 | 0 | 0.5320 | 7.061 | 77.0 | 3.4106 | 24 | 666 | 20.2 | 395.28 | 7.01 | 25.0 |
| 2.81838 | 0.0 | 18.10 | 0 | 0.5320 | 5.762 | 40.3 | 4.0983 | 24 | 666 | 20.2 | 392.92 | 10.42 | 21.8 |
| 2.37857 | 0.0 | 18.10 | 0 | 0.5830 | 5.871 | 41.9 | 3.7240 | 24 | 666 | 20.2 | 370.73 | 13.34 | 20.6 |
| 3.67367 | 0.0 | 18.10 | 0 | 0.5830 | 6.312 | 51.9 | 3.9917 | 24 | 666 | 20.2 | 388.62 | 10.58 | 21.2 |
| 5.69175 | 0.0 | 18.10 | 0 | 0.5830 | 6.114 | 79.8 | 3.5459 | 24 | 666 | 20.2 | 392.68 | 14.98 | 19.1 |
| 4.83567 | 0.0 | 18.10 | 0 | 0.5830 | 5.905 | 53.2 | 3.1523 | 24 | 666 | 20.2 | 388.22 | 11.45 | 20.6 |
| 0.15086 | 0.0 | 27.74 | 0 | 0.6090 | 5.454 | 92.7 | 1.8209 | 4 | 711 | 20.1 | 395.09 | 18.06 | 15.2 |
| 0.18337 | 0.0 | 27.74 | 0 | 0.6090 | 5.414 | 98.3 | 1.7554 | 4 | 711 | 20.1 | 344.05 | 23.97 | 7.0 |
| 0.20746 | 0.0 | 27.74 | 0 | 0.6090 | 5.093 | 98.0 | 1.8226 | 4 | 711 | 20.1 | 318.43 | 29.68 | 8.1 |
| 0.10574 | 0.0 | 27.74 | 0 | 0.6090 | 5.983 | 98.8 | 1.8681 | 4 | 711 | 20.1 | 390.11 | 18.07 | 13.6 |
| 0.11132 | 0.0 | 27.74 | 0 | 0.6090 | 5.983 | 83.5 | 2.1099 | 4 | 711 | 20.1 | 396.90 | 13.35 | 20.1 |
| 0.17331 | 0.0 | 9.69 | 0 | 0.5850 | 5.707 | 54.0 | 2.3817 | 6 | 391 | 19.2 | 396.90 | 12.01 | 21.8 |
| 0.27957 | 0.0 | 9.69 | 0 | 0.5850 | 5.926 | 42.6 | 2.3817 | 6 | 391 | 19.2 | 396.90 | 13.59 | 24.5 |
| 0.17899 | 0.0 | 9.69 | 0 | 0.5850 | 5.670 | 28.8 | 2.7986 | 6 | 391 | 19.2 | 393.29 | 17.60 | 23.1 |
| 0.28960 | 0.0 | 9.69 | 0 | 0.5850 | 5.390 | 72.9 | 2.7986 | 6 | 391 | 19.2 | 396.90 | 21.14 | 19.7 |
| 0.26838 | 0.0 | 9.69 | 0 | 0.5850 | 5.794 | 70.6 | 2.8927 | 6 | 391 | 19.2 | 396.90 | 14.10 | 18.3 |
| 0.23912 | 0.0 | 9.69 | 0 | 0.5850 | 6.019 | 65.3 | 2.4091 | 6 | 391 | 19.2 | 396.90 | 12.92 | 21.2 |
| 0.17783 | 0.0 | 9.69 | 0 | 0.5850 | 5.569 | 73.5 | 2.3999 | 6 | 391 | 19.2 | 395.77 | 15.10 | 17.5 |
| 0.22438 | 0.0 | 9.69 | 0 | 0.5850 | 6.027 | 79.7 | 2.4982 | 6 | 391 | 19.2 | 396.90 | 14.33 | 16.8 |
| 0.06263 | 0.0 | 11.93 | 0 | 0.5730 | 6.593 | 69.1 | 2.4786 | 1 | 273 | 21.0 | 391.99 | 9.67 | 22.4 |
| 0.04527 | 0.0 | 11.93 | 0 | 0.5730 | 6.120 | 76.7 | 2.2875 | 1 | 273 | 21.0 | 396.90 | 9.08 | 20.6 |
| 0.06076 | 0.0 | 11.93 | 0 | 0.5730 | 6.976 | 91.0 | 2.1675 | 1 | 273 | 21.0 | 396.90 | 5.64 | 23.9 |
| 0.10959 | 0.0 | 11.93 | 0 | 0.5730 | 6.794 | 89.3 | 2.3889 | 1 | 273 | 21.0 | 393.45 | 6.48 | 22.0 |
| 0.04741 | 0.0 | 11.93 | 0 | 0.5730 | 6.030 | 80.8 | 2.5050 | 1 | 273 | 21.0 | 396.90 | 7.88 | 11.9 |