43 基于树的方法
43.1 树回归的简单演示
决策树方法按不同自变量的不同值, 分层地把训练集分组。 每层使用一个变量, 所以这样的分组构成一个二叉树表示。 为了预测一个观测的类归属, 找到它所属的组, 用组的类归属或大多数观测的类归属进行预测。 这样的方法称为决策树(decision tree)。 决策树方法既可以用于判别问题, 也可以用于回归问题,称为回归树。
决策树的好处是容易解释, 在自变量为分类变量时没有额外困难。 但预测准确率可能比其它有监督学习方法差。
改进方法包括装袋法(bagging)、随机森林(random forests)、 提升法(boosting)。 这些改进方法都是把许多棵树合并在一起, 通常能改善准确率但是可解释性变差。
对Hitters数据,用Years和Hits作因变量预测log(Salaray)。
## [1] 263 20library(rsample)
set.seed(101)
hit_split <- initial_split(
da_hit, prop = 0.80, strata = Salary)
hit_train <- training(hit_split)
hit_test <- testing(hit_split)在训练集上建立完整的树:
剪枝为只有3个叶结点:
显示树:
## node), split, n, deviance, yval
## * denotes terminal node
##
## 1) root 208 161.20 5.936
## 2) Years < 4.5 72 35.07 5.162 *
## 3) Years > 4.5 136 60.05 6.346
## 6) Hits < 117.5 70 23.60 5.986 *
## 7) Hits > 117.5 66 17.75 6.728 *显示概括:
##
## Regression tree:
## snip.tree(tree = tr1, nodes = c(6L, 2L))
## Number of terminal nodes: 3
## Residual mean deviance: 0.3727 = 76.41 / 205
## Distribution of residuals:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -2.2280 -0.3740 -0.0589 0.0000 0.3414 2.5010做树图:
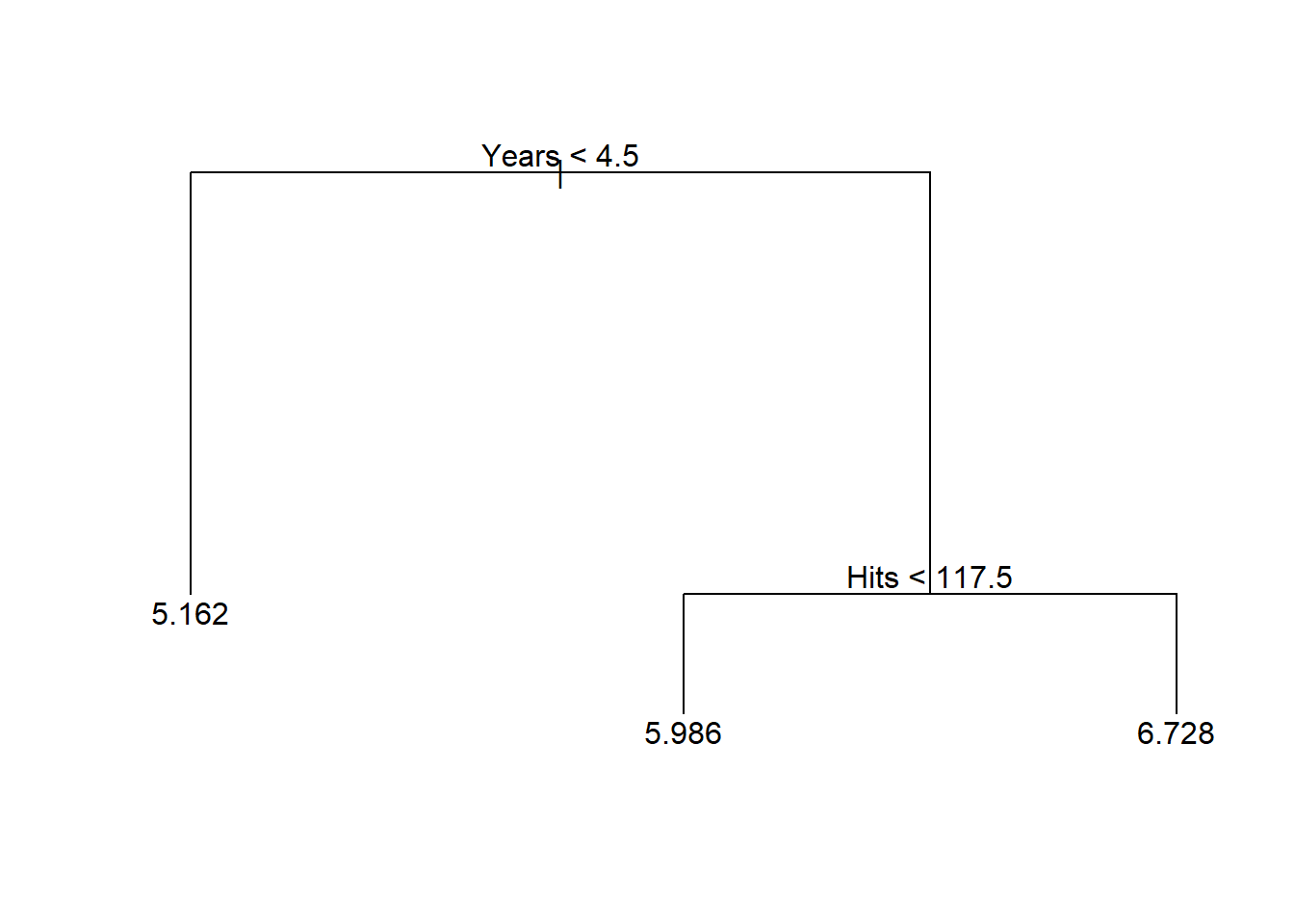
树的深度(depth)是指从根节点到最远的叶节点经过的步数, 比如,上图的树的深度为2, 为了用叶结点给出因变量预测值, 最多需要2次判断。
43.2 树回归
树的深度是一个复杂度指标, 是判别树的超参数, 需要调优。 关于如何进行超参数调优并在测试集上计算性能, tidymodels有系统的方法, 参见47.3。 这里为了对方法进行更直接的演示, 直接调用交叉验证函数进行超参数调优并在测试集上计算预测精度指标。
对训练集上的未剪枝树用交叉验证方法寻找最优大小:
## $size
## [1] 9 8 6 5 4 3 2 1
##
## $dev
## [1] 78.50049 81.47727 81.43670 79.43120 79.07190 92.16026 105.14082 167.75233
##
## $k
## [1] -Inf 2.445601 2.639571 3.186007 4.133744 8.296626 18.711912 66.037022
##
## $method
## [1] "deviance"
##
## attr(,"class")
## [1] "prune" "tree.sequence"plot(cv1$size, cv1$dev, type='b')
best.size <- cv1$size[which.min(cv1$dev)[1]]
abline(v=best.size, col='gray')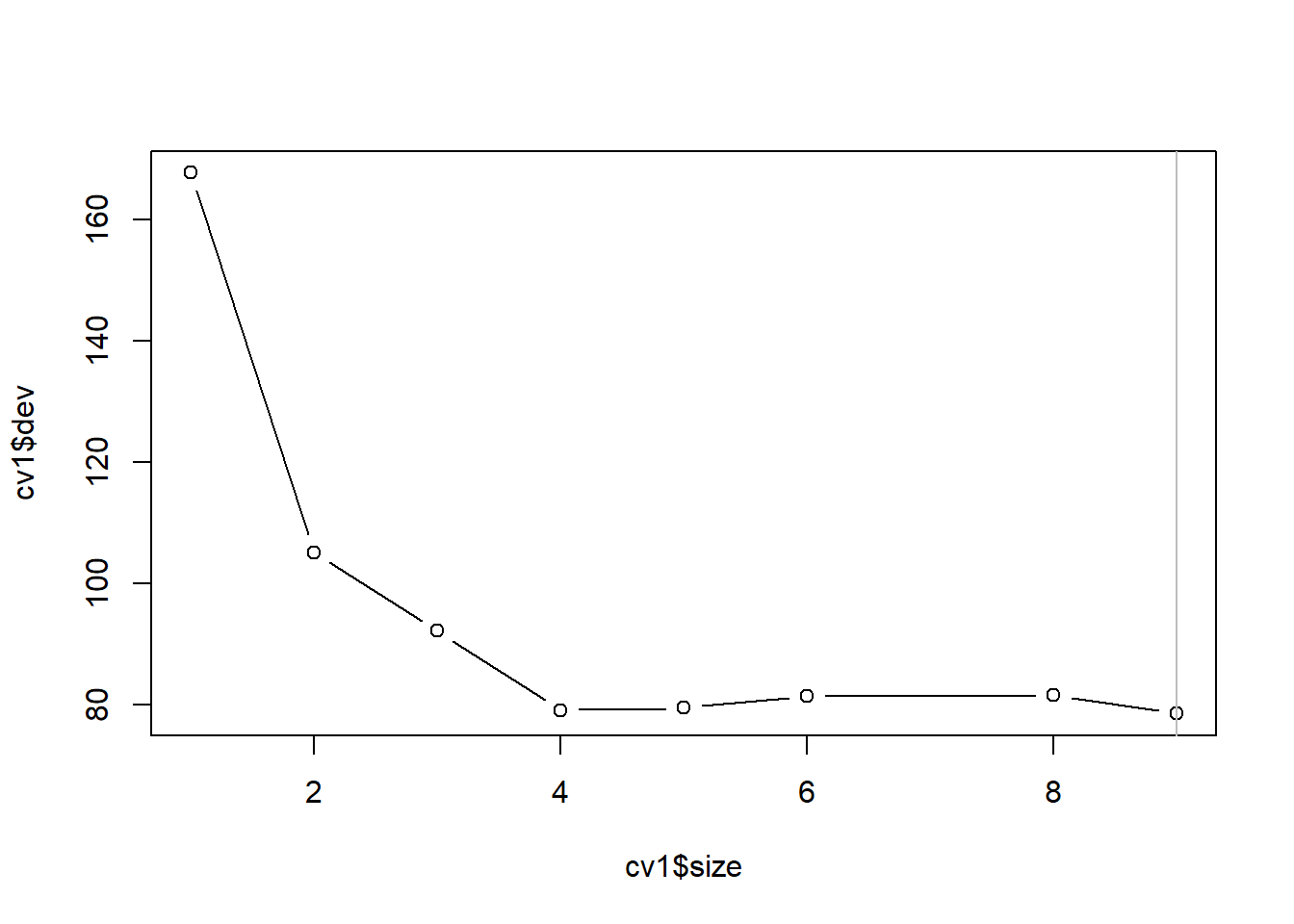
最优大小为9。 但是从图上看, 大小4的树已经效果很好。
获得训练集上构造的树剪枝后的结果:
在测试集上计算预测根均方误差:
pred.test <- predict(tr1b, newdata = hit_test)
test.rmse <-
mean( (hit_test$Salary - exp(pred.test))^2 ) |> sqrt()
test.rmse## [1] 281.7956RMSE=281.8, 比子集回归、岭回归(RMSE=240.7)、lasso的结果都差很多。
用训练集的因变量平均值估计测试集的因变量值可以作为一个最初等的用来对比的基准, 其根均方误差为:
## [1] 413.1353用所有数据来构造未剪枝树:
用训练集上得到的子树大小剪枝:
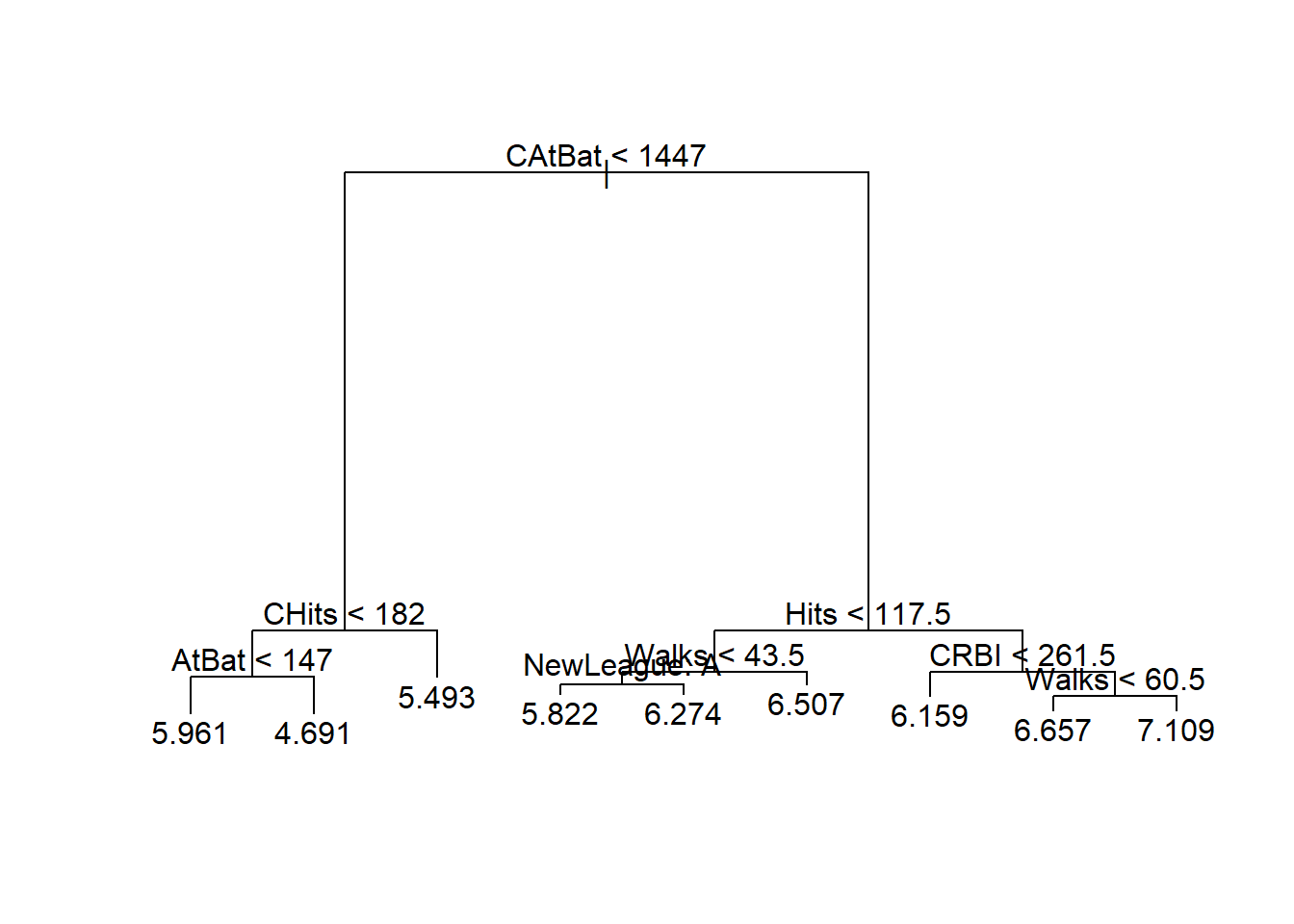
这样的结果可以用于同一问题的新数据的预测。
43.3 装袋法
判别树在不同的训练集、测试集划分上可以产生很大变化, 说明其预测值方差较大。 利用bootstrap的思想, 可以随机选取许多个训练集, 把许多个训练集的模型结果平均, 就可以降低预测值的方差。
办法是从一个训练集中用有放回抽样的方法抽取\(B\)个训练集, 设第\(b\)个抽取的训练集得到的回归函数为\(\hat f^{*b}(\cdot)\), 则最后的回归函数是这些回归函数的平均值: \[\begin{aligned} \hat f_{\text{bagging}}(x) = \frac{1}{B} \sum_{b=1}^b \hat f^{*b}(x) . \end{aligned}\] 这称为装袋法(bagging)。 装袋法对改善判别与回归树的预测精度十分有效。
装袋法的步骤如下:
- 从训练集中取\(B\)个有放回随机抽样的bootstrap训练集,\(B\)取为几百到几千之间。
- 对每个bootstrap训练集,估计未剪枝的树。
- 如果因变量是连续变量,对测试样品,用所有的树的预测值的平均值作预测。
- 如果因变量是分类变量,对测试样品,可以用所有树预测类的多数投票决定预测值。
装袋法也可以用来改进其他的回归和判别方法。
装袋后不能再用图形表示,模型可解释性较差。 但是,可以度量自变量在预测中的重要程度。 在回归问题中, 可以计算每个自变量在所有\(B\)个树中平均减少的残差平方和的量, 以此度量其重要度。 在判别问题中, 可以计算每个自变量在所有\(B\)个树种平均减少的基尼系数的量, 以此度量其重要度。
除了可以用测试集、交叉验证方法, 还可以使用袋外观测的预测误差来度量模型预测精度。 用bootstrap再抽样获得多个训练集时每个bootstrap训练集总会遗漏一些观测, 平均每个bootstrap训练集会遗漏三分之一的观测。 对每个观测,大约有\(B/3\)棵树没有用到此观测, 可以用这些树的预测值平均来预测此观测,得到一个误差估计, 这样得到的均方误差估计或错判率称为袋外观测估计(OOB估计)。 好处是不用很多额外的工作。
对训练集用装袋法:
library(randomForest)
bag1 <- randomForest(
log(Salary) ~ .,
data = hit_train,
mtry=ncol(hit_train)-1,
importance=TRUE)
bag1##
## Call:
## randomForest(formula = log(Salary) ~ ., data = hit_train, mtry = ncol(hit_train) - 1, importance = TRUE)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 19
##
## Mean of squared residuals: 0.1980098
## % Var explained: 74.44注意randomForest()函数实际是随机森林法,
但是当mtry的值取为所有自变量个数时就是装袋法。
对测试集进行预报:
pred2 <- predict(bag1, newdata = hit_test)
test.rmse2 <-
mean( (hit_test$Salary - exp(pred2))^2 ) |> sqrt()
test.rmse2## [1] 202.0765RMSE=202.1, 比判别树的281.8改进很大, 比岭回归的240.7也有很大优势。
在全集上使用装袋法:
##
## Call:
## randomForest(formula = log(Salary) ~ ., data = da_hit, mtry = ncol(da_hit) - 1, importance = TRUE)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 19
##
## Mean of squared residuals: 0.1937377
## % Var explained: 75.4变量的重要度数值和图形: 各变量的重要度数值及其图形:
## %IncMSE IncNodePurity
## AtBat 10.7883286 8.1667778
## Hits 8.4949590 8.1050931
## HmRun 3.0595593 1.9280305
## Runs 7.6675720 3.8182568
## RBI 4.5596220 5.2948207
## Walks 8.0850741 6.9407788
## Years 10.0302334 2.2203968
## CAtBat 26.2359706 77.4088339
## CHits 12.8371027 24.0757798
## CHmRun 4.4959747 4.3641893
## CRuns 14.9272144 36.1514017
## CRBI 15.6525107 11.3891366
## CWalks 6.7160244 6.5333487
## League -0.7821402 0.2073524
## Division -1.0121206 0.2339053
## PutOuts 0.2771301 3.7336895
## Assists -2.5795517 1.7112880
## Errors 0.9658563 1.7447031
## NewLeague 1.2244401 0.3597582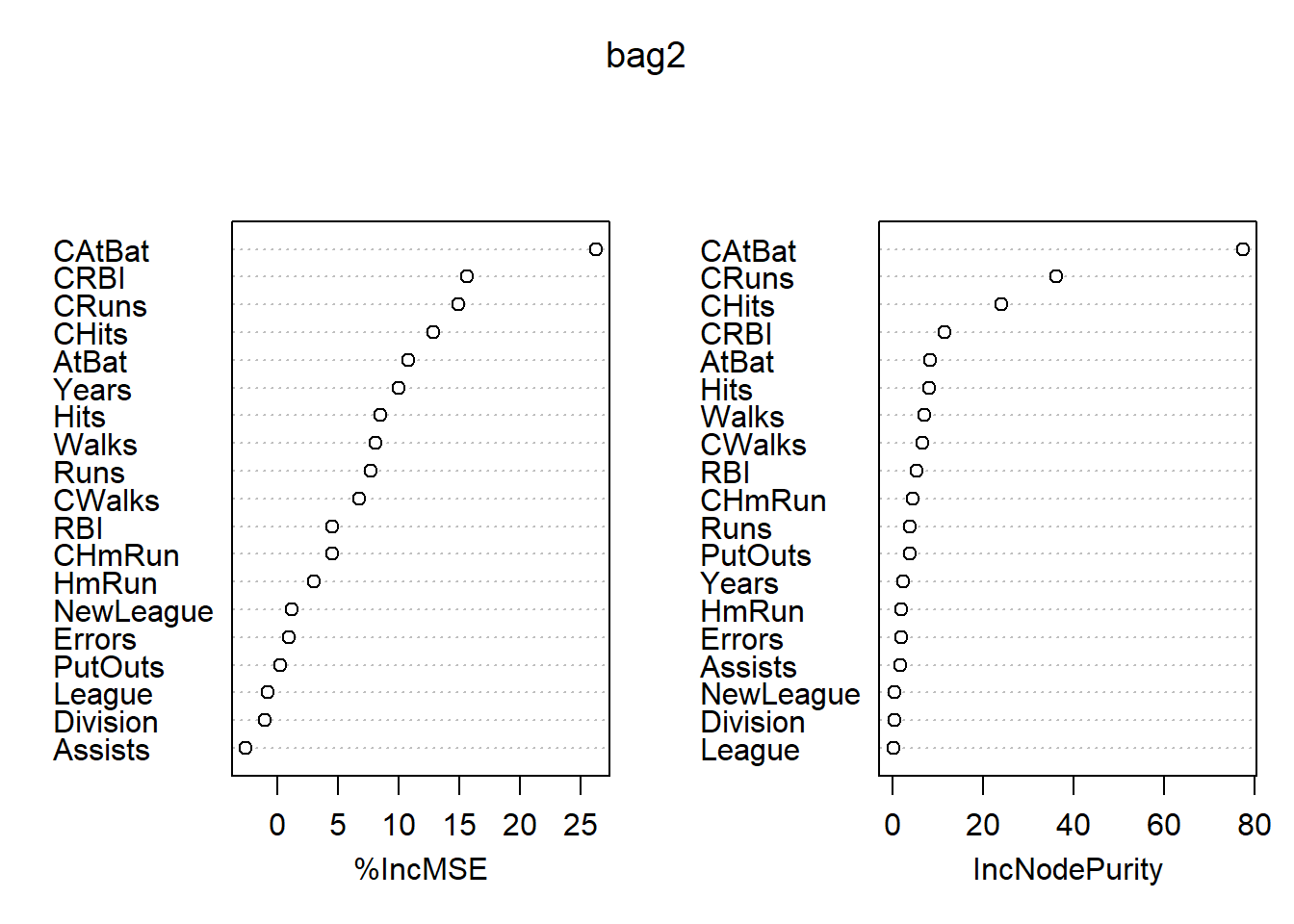
图43.1: Hitters数据装袋法的变量重要性结果
最重要的自变量是CAtBats, 其次有CRuns, CHits等。
如何计算变量重要度? 基于树的方法, 每个叶节点的纯度越高(叶结点中所有观测的标签相同,或者因变量值相等), 模型拟合优度越好。 所以, 对每一个变量, 可以计算其在作为分枝用的变量时, 对中间节点的纯度指标的改善量, 将这些改善量加起来。 对装袋法、随机森林、提升法(如GBM), 则是计算每个变量对损失函数的改善量。
不同的机器学习算法对变量重要程度有不同的定义, 比如, 广义线性模型(GLM)用标准化后的自变量的系数估计的绝对值大小作为重要程度度量。
43.4 随机森林
随机森林的思想与装袋法类似, 但是试图使得参加平均的各个树之间变得比较独立, 以减少正相关的预测在计算平均时的标准误差膨胀问题。 仍采用有放回抽样得到的多个bootstrap训练集, 但是对每个bootstrap训练集构造判别树时, 每次分叉时不考虑所有自变量, 而是仅考虑随机选取的一个自变量子集。 这个自变量子集的自变量个数是一个模型超参数。
对判别树, 每次分叉时选取的自变量个数通常取\(m \approx \sqrt{p}\)个。 比如,对Heart数据的13个自变量, 每次分叉时仅随机选取4个纳入考察范围。
随机森林的想法是基于正相关的样本在平均时并不能很好地降低方差, 独立样本能比较好地降低方差。 如果存在一个最重要的变量, 如果不加限制这个最重要的变量总会是第一个分叉, 使得\(B\)棵树相似程度很高。 随机森林解决这个问题的办法是限制分叉时可选的变量子集。
随机森林也可以用来改进其他的回归和判别方法。
装袋法和随机森林都可以用R扩展包randomForest的
randomForest()函数实现。
当此函数的mtry参数取为自变量个数时,执行的就是装袋法;
mtry取缺省值时,执行随机森林算法。
执行随机森林算法时,
randomForest()函数在回归问题时分叉时考虑的自变量个数取\(m \approx p/3\),
在判别问题时取\(m \approx \sqrt{p}\)。
对训练集用随机森林法:
##
## Call:
## randomForest(formula = log(Salary) ~ ., data = hit_train, importance = TRUE)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 6
##
## Mean of squared residuals: 0.1895383
## % Var explained: 75.54当mtry的值取为缺省值时执行随机森林算法。
对测试集进行预报:
pred3 <- predict(rf1, newdata = hit_test)
test.rmse3 <-
mean( (hit_test$Salary - exp(pred3))^2 ) |> sqrt()
test.rmse3## [1] 199.8305RMSE=199.8, 与装袋法(RMSE=202.1)相近。
在全集上使用随机森林:
##
## Call:
## randomForest(formula = log(Salary) ~ ., data = da_hit, importance = TRUE)
## Type of random forest: regression
## Number of trees: 500
## No. of variables tried at each split: 6
##
## Mean of squared residuals: 0.1799338
## % Var explained: 77.16各变量的重要度数值及其图形:
## %IncMSE IncNodePurity
## AtBat 10.8759999 7.4439449
## Hits 8.1725427 7.9481573
## HmRun 4.4016043 2.5935154
## Runs 9.2818801 4.8293772
## RBI 8.3514919 6.2292463
## Walks 8.8164532 6.1787450
## Years 10.6053647 5.0062719
## CAtBat 16.9507148 41.0814114
## CHits 17.6578387 41.5968368
## CHmRun 8.1292431 7.1035557
## CRuns 13.8588073 30.0948238
## CRBI 14.2775671 19.7903282
## CWalks 10.3261013 15.7222964
## League 2.0932305 0.2700378
## Division -0.2466121 0.3021408
## PutOuts 3.1669627 3.2670212
## Assists -0.6733127 1.7261075
## Errors 1.5649441 1.6376596
## NewLeague 1.0967640 0.3386188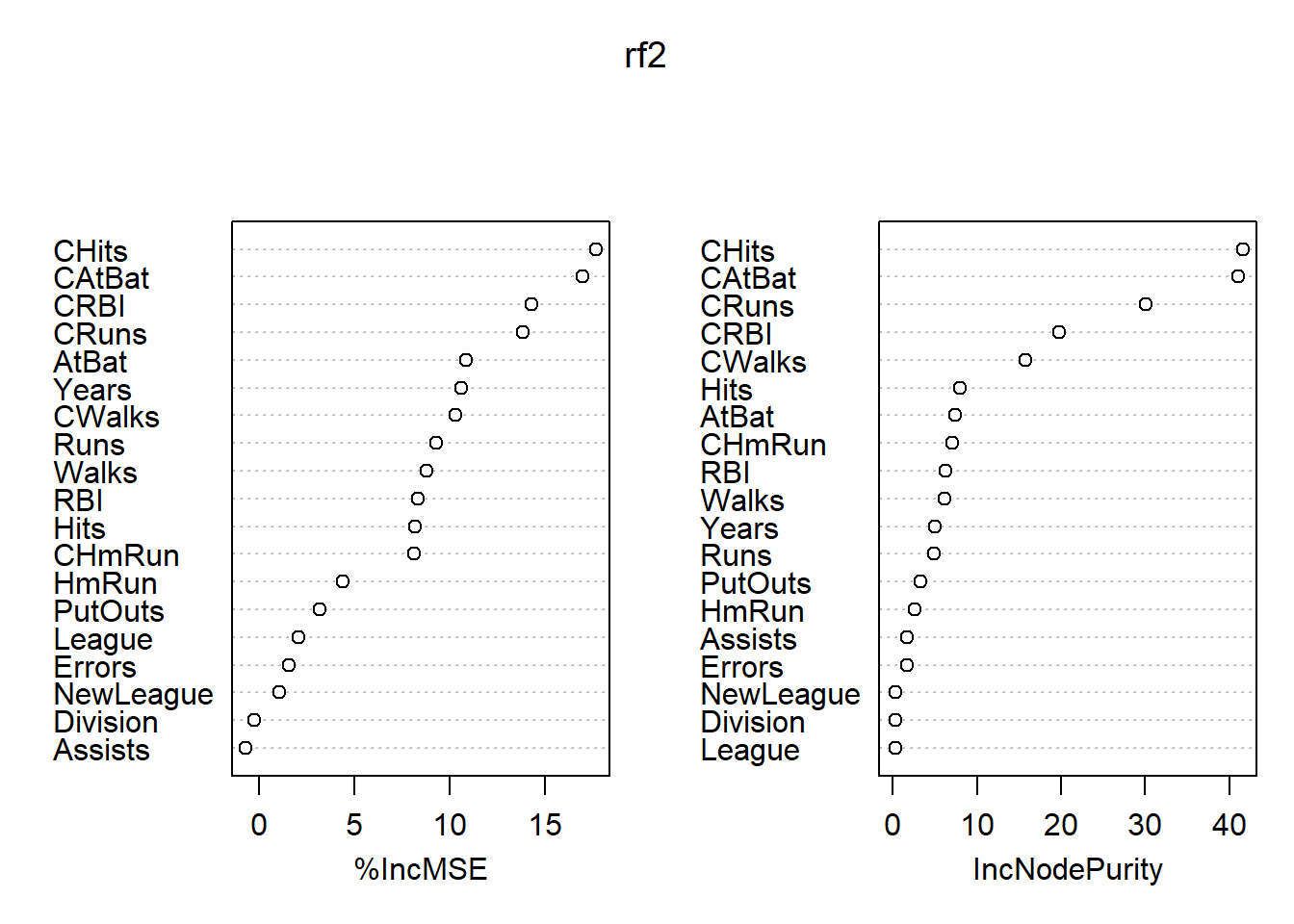
图43.2: Hitters数据随机森林法的变量重要度结果
最重要的自变量是CAtBats, CRuns, CHits, CWalks, CRBI等。
43.5 提升法
提升法(Boosting), 也称为梯度提升法, 也是可以用在多种回归和判别问题中的方法。 提升法的想法是, 用比较简单的模型拟合因变量, 计算残差, 然后以残差为新的因变量建模, 仍使用简单的模型, 把两次的回归函数作加权和, 得到新的残差后,再以新残差作为因变量建模, 如此重复地更新回归函数, 得到由多个回归函数加权和组成的最终的回归函数。
加权一般取为比较小的值, 其目的是降低逼近速度。 统计学习问题中降低逼近速度一般结果更好。
提升法算法:
[(1)] 对训练集,设置\(r_i = y_i\),并令初始回归函数为\(\hat f(\cdot)=0\)。
[(2)] 对\(b=1,2,\dots,B\)重复执行:
- [(a)] 以训练集的自变量为自变量,以\(r\)为因变量,拟合一个仅有\(d\)个分叉的简单树回归函数, 设为\(\hat f_b\);
- [(b)] 更新回归函数,添加一个压缩过的树回归函数: \[\begin{aligned} \hat f(x) \leftarrow \hat f(x) + \lambda \hat f_b(x); \end{aligned}\]
- [(c)] 更新残差: \[\begin{aligned} r_i \leftarrow r_i - \lambda \hat f_b(x_i). \end{aligned}\]
[(3)] 提升法的回归函数为 \[\begin{aligned} \hat f(x) = \sum_{b=1}^B \lambda \hat f_b(x) . \end{aligned}\]
用多少个回归函数做加权和,即\(B\)的选取问题。 取得\(B\)太大也会有过度拟合, 但是只要\(B\)不太大这个问题不严重。 可以用交叉验证选择\(B\)的值。
收缩系数\(\lambda\)。 是一个小的正数, 控制学习速度, 经常用0.01, 0.001这样的值, 与要解决的问题有关。 取\(\lambda\)很小,就需要取\(B\)很大。
用来控制每个回归函数复杂度的参数, 对树回归而言就是树的大小, 用树的深度\(d\)表示。 深度等于1则仅使用一个自变量, 仅有一次分叉, 就是二叉树, 这样多棵树相加, 相当于各个变量的可加模型, 没有交互作用效应, 这样的可加模型往往就很好。 \(d>1\)时就加入了交互项, 比如\(d=2\), 就可以用两个变量, 用叶结点预测因变量时, 最多可以用两个自变量作两次判断, 因为树模型是非线性的, 将许多棵这样的深度为2的树相加, 就可以包含自变量两两之间的非线性的相互作用效应。
gbm实现了提升法。
interaction.depth表示树的深度(复杂度),
n.trees表示用多少棵树相加。
shrinkage表示学习速度,
即算法中的\(\lambda\)。
n.minobsinnode表示每个叶结点至少应包含的观测点数,
可以设置这个参数,
以避免过少的训练样例也单独作为一个规则。
这些都是超参数,
应进行超参数调优,
这里仅固定了这些超参数进行演示。
在训练集上拟合:
library(gbm)
set.seed(1)
bst1 <- gbm(
log(Salary) ~ .,
data = hit_train,
distribution = "gaussian",
n.trees=5000,
interaction.depth=4)
summary(bst1)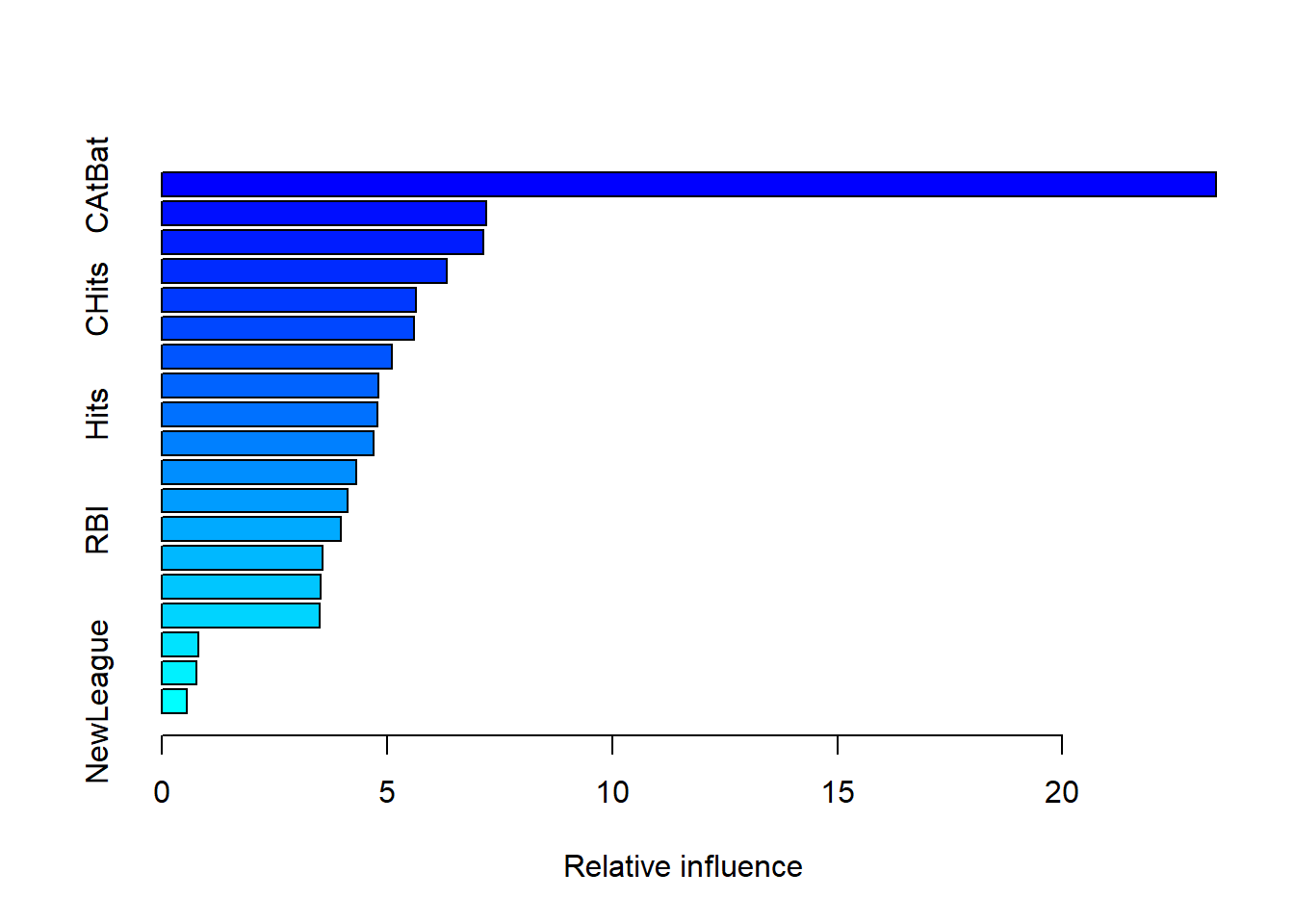
## var rel.inf
## CAtBat CAtBat 23.4075576
## CRBI CRBI 7.2138130
## CRuns CRuns 7.1524081
## PutOuts PutOuts 6.3402558
## CHits CHits 5.6558782
## CHmRun CHmRun 5.6051624
## Walks Walks 5.1110904
## Assists Assists 4.8197073
## Hits Hits 4.7970012
## CWalks CWalks 4.7150910
## AtBat AtBat 4.3214885
## HmRun HmRun 4.1297511
## RBI RBI 3.9799787
## Years Years 3.5699618
## Runs Runs 3.5257357
## Errors Errors 3.5019377
## Division Division 0.8191874
## League League 0.7703509
## NewLeague NewLeague 0.5636432CAtBat是最重要的变量。
在测试集上预报,并计算根均方误差:
## Using 5000 trees...## [1] 274.633RMSE=274.6, 结果比较差, 需要进行参数调优。
43.6 心脏病诊断建模预报
Heart数据是心脏病诊断的数据, 因变量AHD为是否有心脏病, 试图用各个自变量预测(判别)。
读入Heart数据集,并去掉有缺失值的观测:
Heart <- read_csv(
"data/Heart.csv",
show_col_types = FALSE) |>
dplyr::select(-1) |>
mutate(
AHD = factor(AHD, levels=c("Yes", "No"))
)## New names:
## • `` -> `...1`## Rows: 297
## Columns: 14
## $ Age <dbl> 63, 67, 67, 37, 41, 56, 62, 57, 63, 53, 57, 56, 56, 44, 52, 57, 48, 54, 48, 49, 64, 58, 58, 58, 60, 50, 58, 66, 43, 40, 69, 60, 64, 59, 44, 42, 43, 57, 55, 61, 65, 40, 71, 59, 61, 58, 51, 50, 65, 53, 41, 65, 44, 44, 60, 54, 50, 41, 54, 51, 51, 46, 58, 54, 54, 60, 60, 54, 59, 46, 65, 67, 62, 65, 44, 65, 60, 51, 48, 58, 45, 53, 39, 68, 52, 44, 47, 53, 51, 66, 62, 62, 44, 63, 52, 59, 60, 52, 48, 45, 34, 57, 71, 49, 54, 59, 57, 61, 39, 61, 56, 52, 43, 62, 41, 58, 35, 63, 65, 48, …
## $ Sex <dbl> 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 0, 0, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 0, 0, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 0, 0, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, …
## $ ChestPain <chr> "typical", "asymptomatic", "asymptomatic", "nonanginal", "nontypical", "nontypical", "asymptomatic", "asymptomatic", "asymptomatic", "asymptomatic", "asymptomatic", "nontypical", "nonanginal", "nontypical", "nonanginal", "nonanginal", "nontypical", "asymptomatic", "nonanginal", "nontypical", "typical", "typical", "nontypical", "nonanginal", "asymptomatic", "nonanginal", "nonanginal", "typical", "asymptomatic", "asymptomatic", "typical", "asymptomatic", "nonanginal", "asymptom…
## $ RestBP <dbl> 145, 160, 120, 130, 130, 120, 140, 120, 130, 140, 140, 140, 130, 120, 172, 150, 110, 140, 130, 130, 110, 150, 120, 132, 130, 120, 120, 150, 150, 110, 140, 117, 140, 135, 130, 140, 120, 150, 132, 150, 150, 140, 160, 150, 130, 112, 110, 150, 140, 130, 105, 120, 112, 130, 130, 124, 140, 110, 125, 125, 130, 142, 128, 135, 120, 145, 140, 150, 170, 150, 155, 125, 120, 110, 110, 160, 125, 140, 130, 150, 104, 130, 140, 180, 120, 140, 138, 138, 130, 120, 160, 130, 108, 135, 128, 110, …
## $ Chol <dbl> 233, 286, 229, 250, 204, 236, 268, 354, 254, 203, 192, 294, 256, 263, 199, 168, 229, 239, 275, 266, 211, 283, 284, 224, 206, 219, 340, 226, 247, 167, 239, 230, 335, 234, 233, 226, 177, 276, 353, 243, 225, 199, 302, 212, 330, 230, 175, 243, 417, 197, 198, 177, 290, 219, 253, 266, 233, 172, 273, 213, 305, 177, 216, 304, 188, 282, 185, 232, 326, 231, 269, 254, 267, 248, 197, 360, 258, 308, 245, 270, 208, 264, 321, 274, 325, 235, 257, 234, 256, 302, 164, 231, 141, 252, 255, 239, …
## $ Fbs <dbl> 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, …
## $ RestECG <dbl> 2, 2, 2, 0, 2, 0, 2, 0, 2, 2, 0, 2, 2, 0, 0, 0, 0, 0, 0, 0, 2, 2, 2, 2, 2, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 2, 2, 0, 0, 2, 0, 0, 0, 2, 2, 0, 2, 2, 2, 0, 0, 2, 2, 0, 2, 0, 2, 2, 2, 0, 2, 2, 0, 0, 2, 2, 2, 2, 0, 0, 0, 0, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 0, 2, 2, 2, 2, 2, 2, 0, 0, 2, 0, 2, 2, 0, 2, 2, 2, 2, 2, 0, 0, 0, 2, 0, 0, 2, 2, 2, 2, 0, 0, 2, 0, 2, 2, 2, 2, 0, 0, 2, 2, 2, 0, 0, 0, 2, 0, 2, 2, 0, 2, 0, 2, 0, 2, 0, 2, 0, 0, 2, 0, 2, 0, 2, 0, 0, 2, 2, 2, 2, 2, 0, 2, 2, 0, 0, …
## $ MaxHR <dbl> 150, 108, 129, 187, 172, 178, 160, 163, 147, 155, 148, 153, 142, 173, 162, 174, 168, 160, 139, 171, 144, 162, 160, 173, 132, 158, 172, 114, 171, 114, 151, 160, 158, 161, 179, 178, 120, 112, 132, 137, 114, 178, 162, 157, 169, 165, 123, 128, 157, 152, 168, 140, 153, 188, 144, 109, 163, 158, 152, 125, 142, 160, 131, 170, 113, 142, 155, 165, 140, 147, 148, 163, 99, 158, 177, 151, 141, 142, 180, 111, 148, 143, 182, 150, 172, 180, 156, 160, 149, 151, 145, 146, 175, 172, 161, 142, 1…
## $ ExAng <dbl> 0, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, …
## $ Oldpeak <dbl> 2.3, 1.5, 2.6, 3.5, 1.4, 0.8, 3.6, 0.6, 1.4, 3.1, 0.4, 1.3, 0.6, 0.0, 0.5, 1.6, 1.0, 1.2, 0.2, 0.6, 1.8, 1.0, 1.8, 3.2, 2.4, 1.6, 0.0, 2.6, 1.5, 2.0, 1.8, 1.4, 0.0, 0.5, 0.4, 0.0, 2.5, 0.6, 1.2, 1.0, 1.0, 1.4, 0.4, 1.6, 0.0, 2.5, 0.6, 2.6, 0.8, 1.2, 0.0, 0.4, 0.0, 0.0, 1.4, 2.2, 0.6, 0.0, 0.5, 1.4, 1.2, 1.4, 2.2, 0.0, 1.4, 2.8, 3.0, 1.6, 3.4, 3.6, 0.8, 0.2, 1.8, 0.6, 0.0, 0.8, 2.8, 1.5, 0.2, 0.8, 3.0, 0.4, 0.0, 1.6, 0.2, 0.0, 0.0, 0.0, 0.5, 0.4, 6.2, 1.8, 0.6, 0.0, 0.0, 1.2, …
## $ Slope <dbl> 3, 2, 2, 3, 1, 1, 3, 1, 2, 3, 2, 2, 2, 1, 1, 1, 3, 1, 1, 1, 2, 1, 2, 1, 2, 2, 1, 3, 1, 2, 1, 1, 1, 2, 1, 1, 2, 2, 2, 2, 2, 1, 1, 1, 1, 2, 1, 2, 1, 3, 1, 1, 1, 1, 1, 2, 2, 1, 3, 1, 2, 3, 2, 1, 2, 2, 2, 1, 3, 2, 1, 2, 2, 1, 1, 1, 2, 1, 2, 1, 2, 2, 1, 2, 1, 1, 1, 1, 1, 2, 3, 2, 2, 1, 1, 2, 2, 1, 1, 1, 1, 1, 1, 2, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 2, 1, 2, 2, 3, 2, 2, 3, 2, 1, 1, 2, 1, 1, 1, 2, 2, 3, 2, 2, 2, 1, 2, 1, 2, 2, 1, 2, 1, 1, 1, 2, 2, 2, 2, 3, 2, 1, 1, 2, 1, 1, …
## $ Ca <dbl> 0, 3, 2, 0, 0, 0, 2, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 2, 2, 0, 0, 0, 0, 0, 2, 2, 0, 0, 0, 0, 0, 1, 1, 0, 3, 0, 2, 0, 0, 1, 0, 0, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 3, 0, 1, 2, 0, 0, 0, 0, 0, 2, 2, 2, 1, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 3, 3, 0, 0, 1, 1, 2, 1, 0, 0, 0, 1, 1, 3, 0, 1, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 3, 1, 2, 3, 0, 0, 1, 0, 2, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0, 1, 0, 0, 0, 1, 1, 3, 0, 2, 2, 1, 0, …
## $ Thal <chr> "fixed", "normal", "reversable", "normal", "normal", "normal", "normal", "normal", "reversable", "reversable", "fixed", "normal", "fixed", "reversable", "reversable", "normal", "reversable", "normal", "normal", "normal", "normal", "normal", "normal", "reversable", "reversable", "normal", "normal", "normal", "normal", "reversable", "normal", "reversable", "normal", "reversable", "normal", "normal", "reversable", "fixed", "reversable", "normal", "reversable", "reversable", "nor…
## $ AHD <fct> No, Yes, Yes, No, No, No, Yes, No, Yes, Yes, No, No, Yes, No, No, No, Yes, No, No, No, No, No, Yes, Yes, Yes, No, No, No, No, Yes, No, Yes, Yes, No, No, No, Yes, Yes, Yes, No, Yes, No, No, No, Yes, Yes, No, Yes, No, No, No, No, Yes, No, Yes, Yes, Yes, Yes, No, No, Yes, No, Yes, No, Yes, Yes, Yes, No, Yes, Yes, No, Yes, Yes, Yes, Yes, No, Yes, No, No, Yes, No, No, No, Yes, No, No, No, No, No, No, Yes, No, No, No, Yes, Yes, Yes, No, No, No, No, No, No, Yes, No, Yes, Yes, Yes, Y…##
## Age Min. :29.00 1st Qu.:48.00 Median :56.00 Mean :54.54 3rd Qu.:61.00 Max. :77.00
## Sex Min. :0.0000 1st Qu.:0.0000 Median :1.0000 Mean :0.6768 3rd Qu.:1.0000 Max. :1.0000
## ChestPain Length:297 Class :character Mode :character
## RestBP Min. : 94.0 1st Qu.:120.0 Median :130.0 Mean :131.7 3rd Qu.:140.0 Max. :200.0
## Chol Min. :126.0 1st Qu.:211.0 Median :243.0 Mean :247.4 3rd Qu.:276.0 Max. :564.0
## Fbs Min. :0.0000 1st Qu.:0.0000 Median :0.0000 Mean :0.1448 3rd Qu.:0.0000 Max. :1.0000
## RestECG Min. :0.0000 1st Qu.:0.0000 Median :1.0000 Mean :0.9966 3rd Qu.:2.0000 Max. :2.0000
## MaxHR Min. : 71.0 1st Qu.:133.0 Median :153.0 Mean :149.6 3rd Qu.:166.0 Max. :202.0
## ExAng Min. :0.0000 1st Qu.:0.0000 Median :0.0000 Mean :0.3266 3rd Qu.:1.0000 Max. :1.0000
## Oldpeak Min. :0.000 1st Qu.:0.000 Median :0.800 Mean :1.056 3rd Qu.:1.600 Max. :6.200
## Slope Min. :1.000 1st Qu.:1.000 Median :2.000 Mean :1.603 3rd Qu.:2.000 Max. :3.000
## Ca Min. :0.0000 1st Qu.:0.0000 Median :0.0000 Mean :0.6768 3rd Qu.:1.0000 Max. :3.0000
## Thal Length:297 Class :character Mode :character
## AHD Yes:137 No :160数据下载:Heart.csv
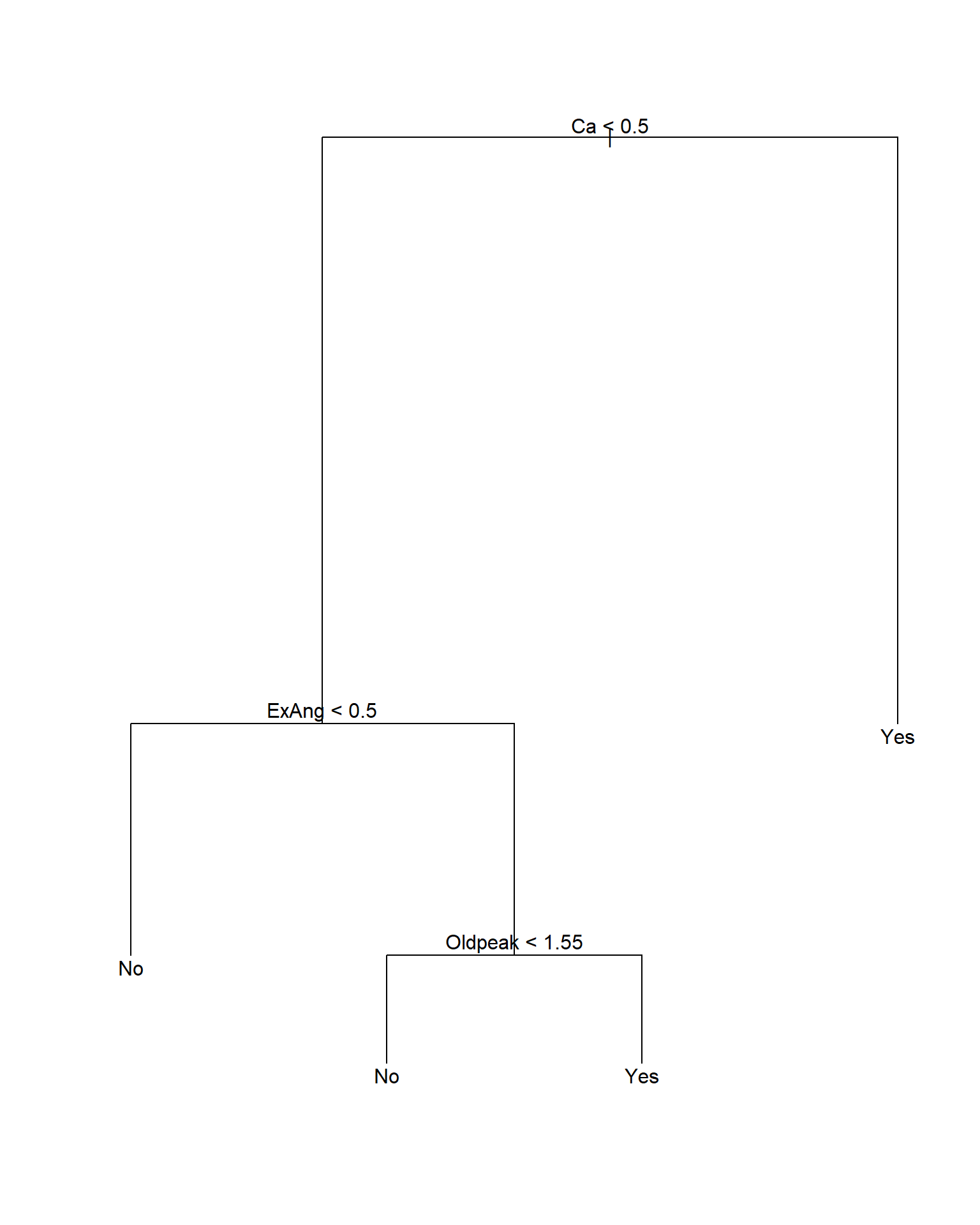
43.6.2 判别树
在训练集上建立未剪枝的判别树:
## Warning in tree(AHD ~ ., data = heart_train): NAs introduced by coercion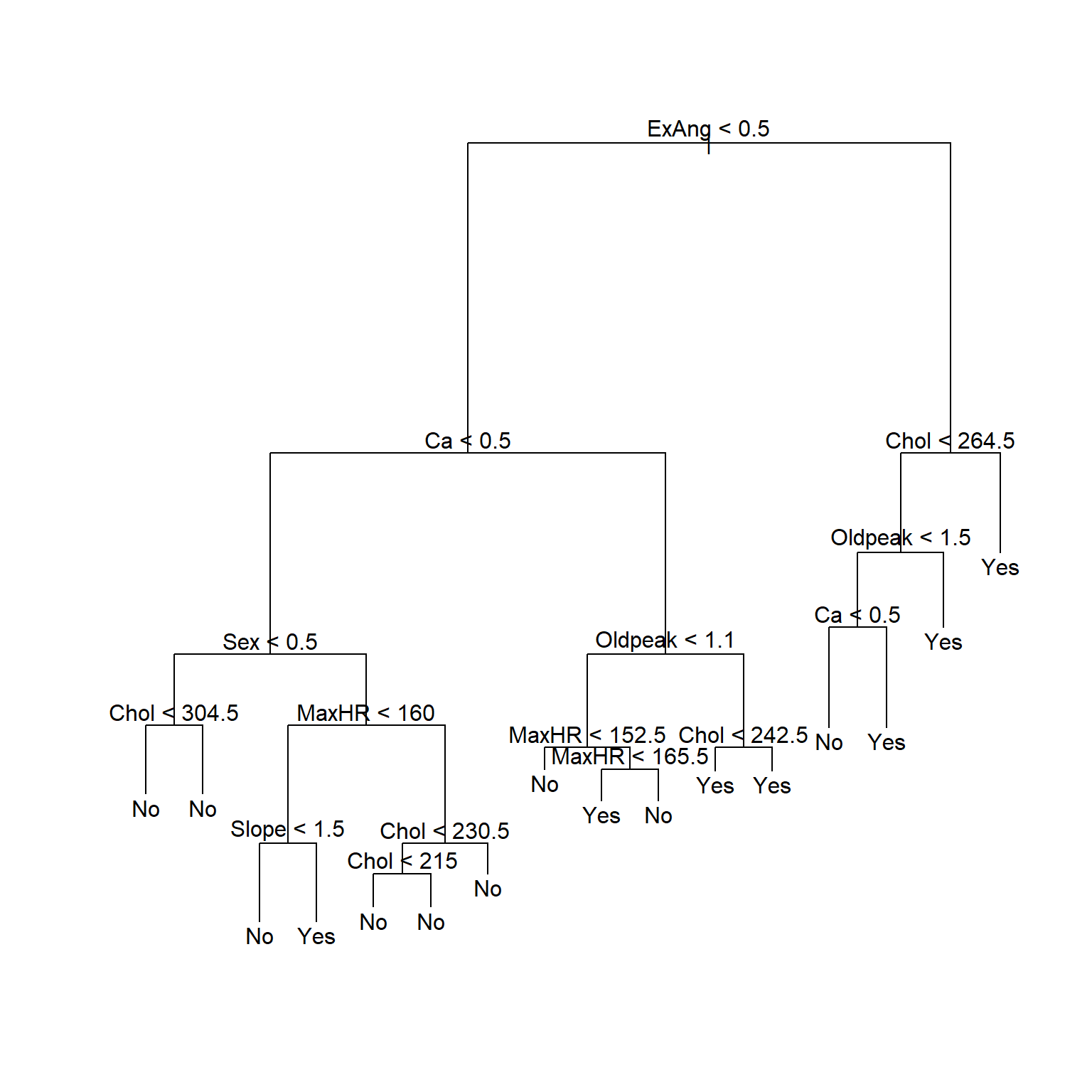
注意剪枝后树的显示中, 如果内部节点的自变量存在分类变量, 这时按照这个自变量分叉时, 取指定的某几个分类值时对应分支Yes, 取其它的分类值时对应分支No。
43.6.2.1 适当剪枝
用交叉验证方法确定剪枝保留的叶子个数, 剪枝时按照错判率(等于1减去正确率)执行:
## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## Warning in tree(model = m[rand != i, , drop = FALSE]): NAs introduced by coercion## Warning in pred1.tree(tree, tree.matrix(nd)): NAs introduced by coercion## $size
## [1] 16 12 6 3 2 1
##
## $dev
## [1] 51 50 53 47 57 75
##
## $k
## [1] -Inf 0.000000 1.666667 2.000000 12.000000 24.000000
##
## $method
## [1] "misclass"
##
## attr(,"class")
## [1] "prune" "tree.sequence"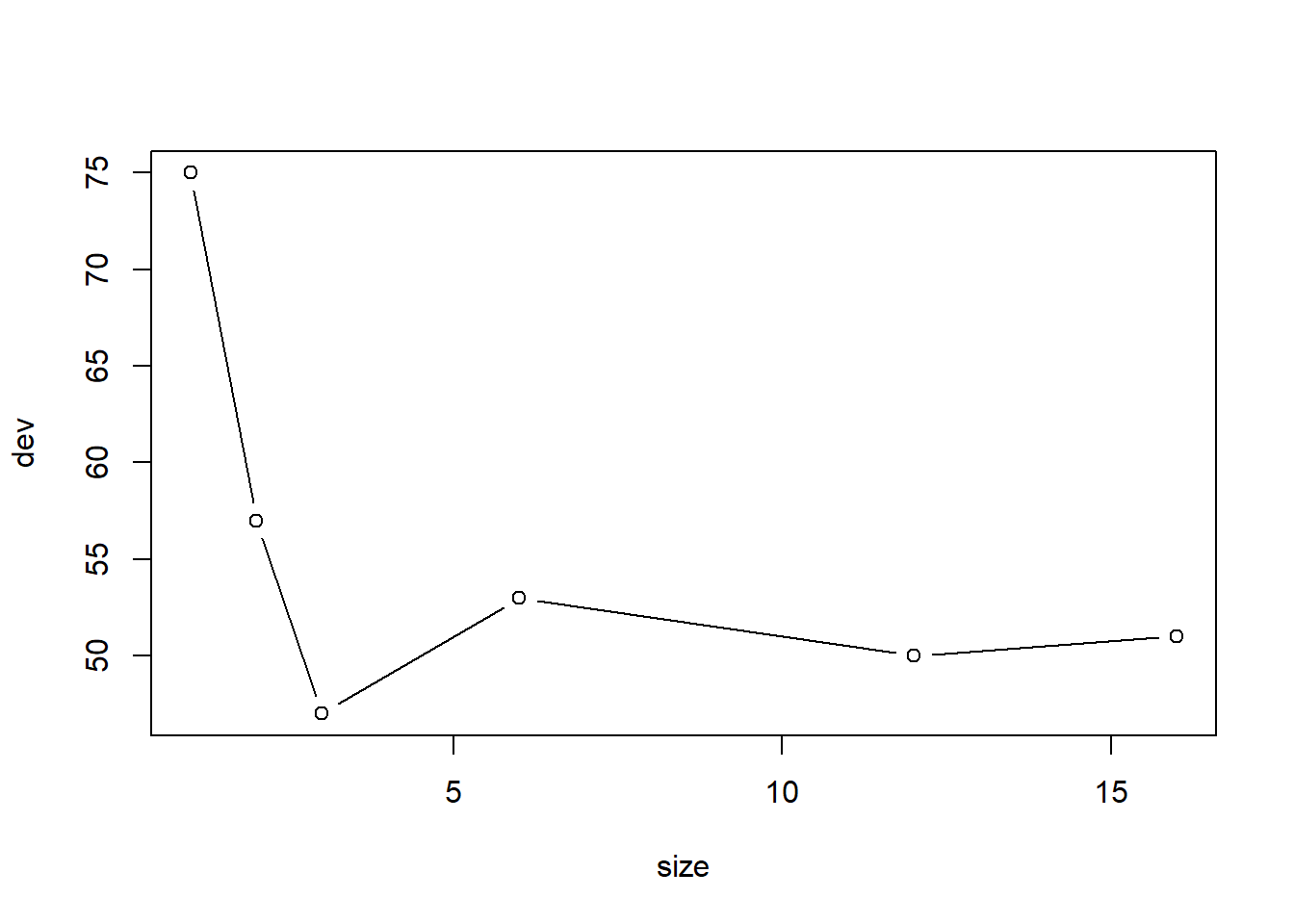
最优的大小是3。
对训练集生成剪枝结果:
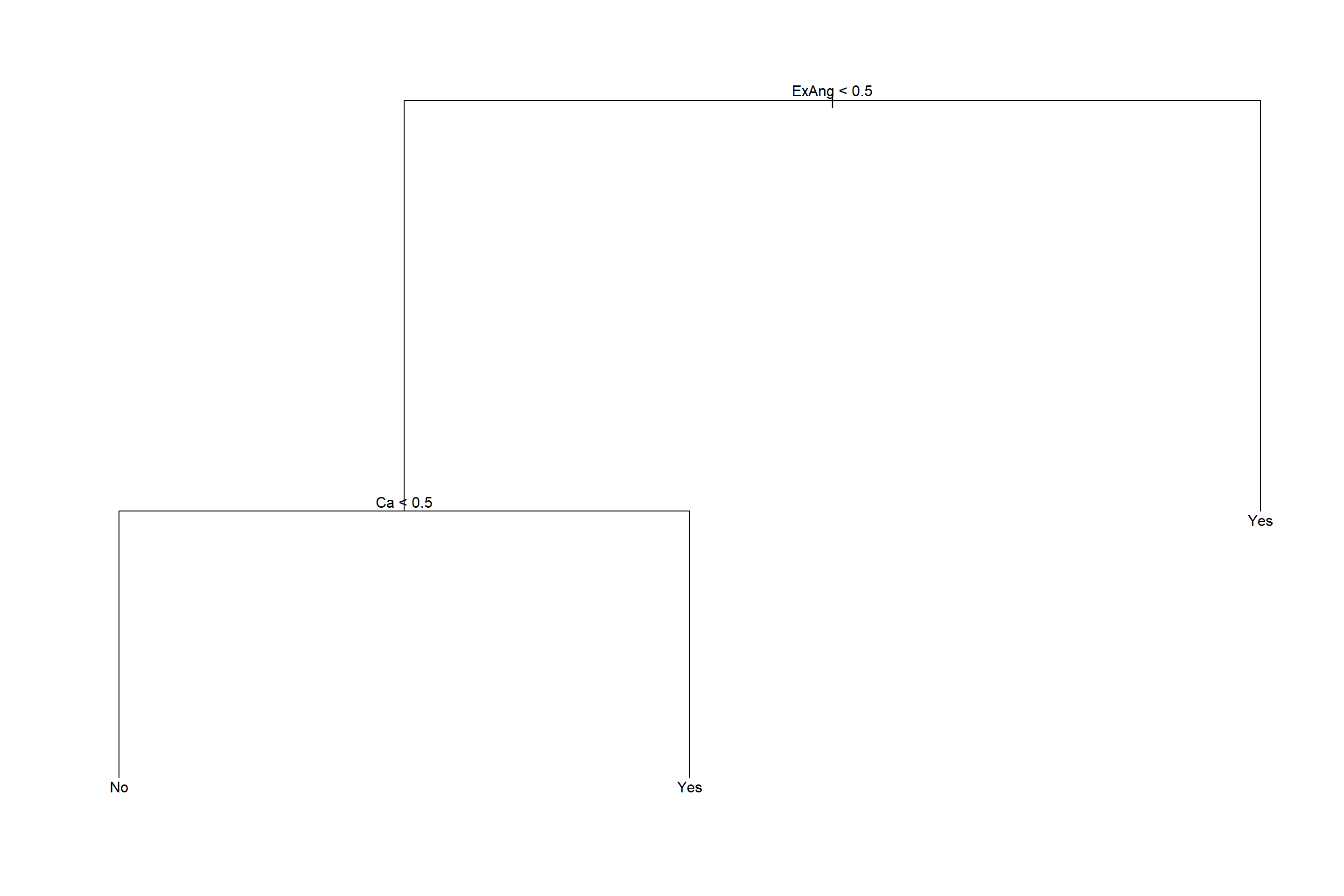
图43.3: Heart数据回归树
43.6.2.2 对测试集计算误判率
## Warning in pred1.tree(object, tree.matrix(newdata)): NAs introduced by coercion## test.y
## pred1 Yes No
## Yes 52 30
## No 6 61## [1] 0.2416107对测试集的错判率约24%。
利用未剪枝的树对测试集进行预测, 一般比剪枝后的结果差:
## Warning in pred1.tree(object, tree.matrix(newdata)): NAs introduced by coercion## test.y
## pred1a Yes No
## Yes 42 25
## No 16 66## [1] 0.275167843.6.3 用装袋法
对训练集用装袋法:
##
## Call:
## randomForest(formula = AHD ~ ., data = heart_train, mtry = 13, importance = TRUE)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 13
##
## OOB estimate of error rate: 23.65%
## Confusion matrix:
## Yes No class.error
## Yes 61 18 0.2278481
## No 17 52 0.2463768注意randomForest()函数实际是随机森林法,
但是当mtry的值取为所有自变量个数时就是装袋法。
袋外观测得到的错判率比较差。
对测试集进行预报:
## test.y
## pred2 Yes No
## Yes 44 15
## No 14 76## [1] 0.1946309测试集的错判率约为19%。
对全集用装袋法:
##
## Call:
## randomForest(formula = AHD ~ ., data = Heart, mtry = 13, importance = TRUE)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 13
##
## OOB estimate of error rate: 21.21%
## Confusion matrix:
## Yes No class.error
## Yes 100 37 0.270073
## No 26 134 0.162500各变量的重要度数值及其图形:
## Yes No MeanDecreaseAccuracy MeanDecreaseGini
## Age 3.7368005 5.9999867 7.2128941 12.1304971
## Sex 8.4082243 11.6854290 14.2202966 4.6253056
## ChestPain 19.2727462 13.7357878 23.2572070 27.9651052
## RestBP 0.1959288 4.4080821 3.5491959 9.7198756
## Chol -4.4853304 1.6204588 -1.8363588 11.5630931
## Fbs -0.9582635 0.5261395 -0.3205312 0.8449148
## RestECG 1.6353427 0.1595635 1.3271811 1.6408234
## MaxHR 2.0318500 8.1264705 7.6843512 13.1458248
## ExAng 5.7030645 1.7850807 5.8043107 3.5962657
## Oldpeak 14.6213004 14.0934301 20.1179974 15.5059594
## Slope 5.6872206 3.5781749 6.3034330 5.5141310
## Ca 18.5244259 25.2918527 30.3846628 22.4696501
## Thal 13.6455796 17.5096952 20.9420003 18.4300152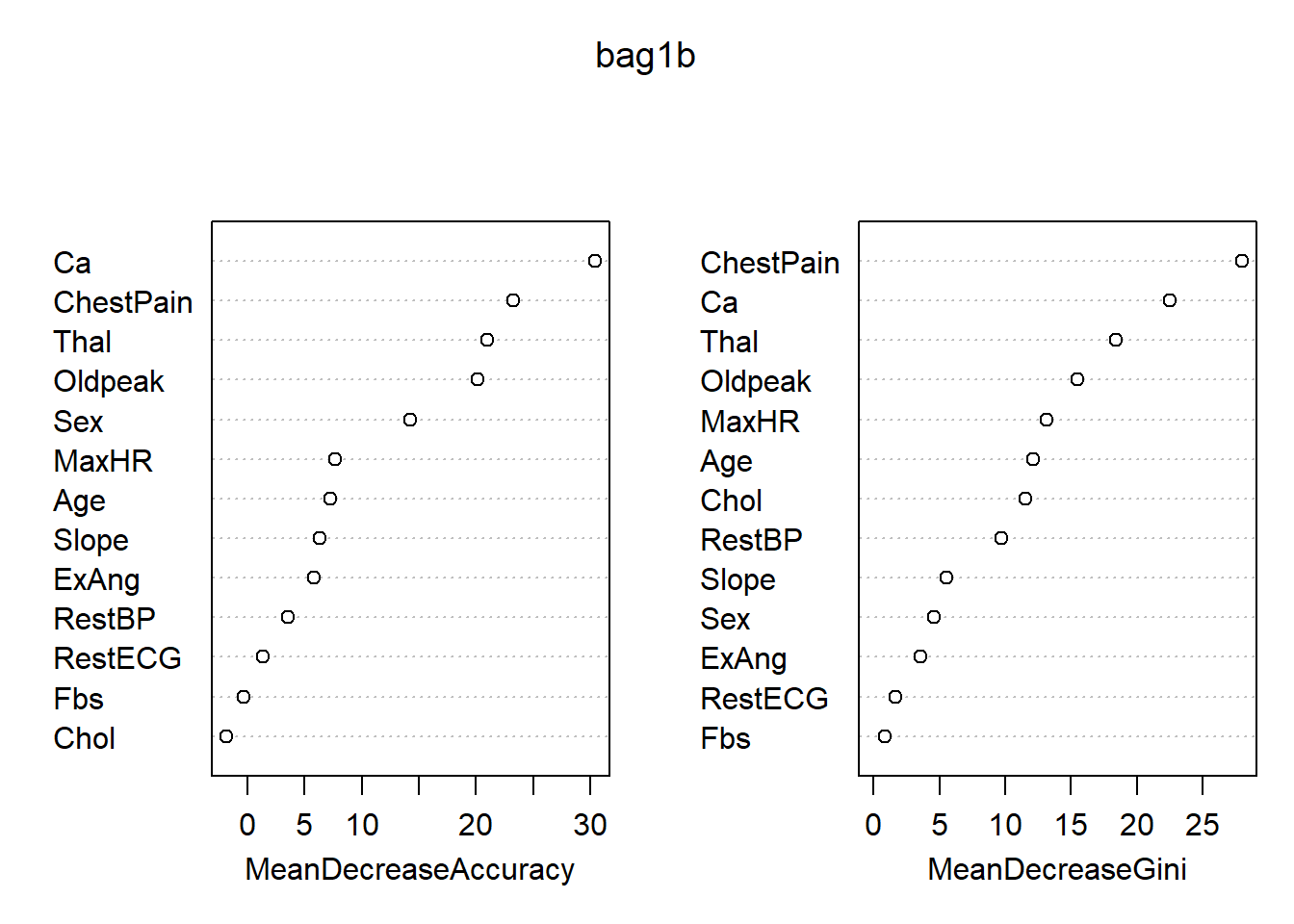
最重要的变量是Thal, ChestPain, Ca。
43.6.4 用随机森林
对训练集用随机森林法:
##
## Call:
## randomForest(formula = AHD ~ ., data = heart_train, importance = TRUE)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 3
##
## OOB estimate of error rate: 22.3%
## Confusion matrix:
## Yes No class.error
## Yes 65 14 0.1772152
## No 19 50 0.2753623这里mtry取缺省值,对应于随机森林法。
对测试集进行预报:
## test.y
## pred3 Yes No
## Yes 47 15
## No 11 76## [1] 0.1744966测试集的错判率约为17%。
对全集用随机森林:
##
## Call:
## randomForest(formula = AHD ~ ., data = Heart, importance = TRUE)
## Type of random forest: classification
## Number of trees: 500
## No. of variables tried at each split: 3
##
## OOB estimate of error rate: 18.86%
## Confusion matrix:
## Yes No class.error
## Yes 108 29 0.2116788
## No 27 133 0.1687500各变量的重要度数值及其图形:
## Yes No MeanDecreaseAccuracy MeanDecreaseGini
## Age 4.164290 6.96750571 8.73313795 12.987241
## Sex 7.281438 10.61442721 13.05548914 4.987433
## ChestPain 17.452527 11.48126517 19.04415445 18.341807
## RestBP 0.190149 1.37058415 1.24243793 10.526087
## Chol -2.275121 1.94668809 -0.03619012 11.468039
## Fbs -1.664923 2.79680029 0.80530655 1.553999
## RestECG 4.645072 -0.02136297 3.19137194 2.815654
## MaxHR 6.711873 8.57257441 10.49071059 17.820488
## ExAng 9.259580 4.60805172 9.61466086 6.847263
## Oldpeak 13.871594 9.53000298 16.91663034 16.497308
## Slope 8.358886 3.06829267 8.36900777 6.832079
## Ca 18.797958 21.81004086 26.74441512 18.984673
## Thal 14.410041 18.71166223 21.30209724 15.882196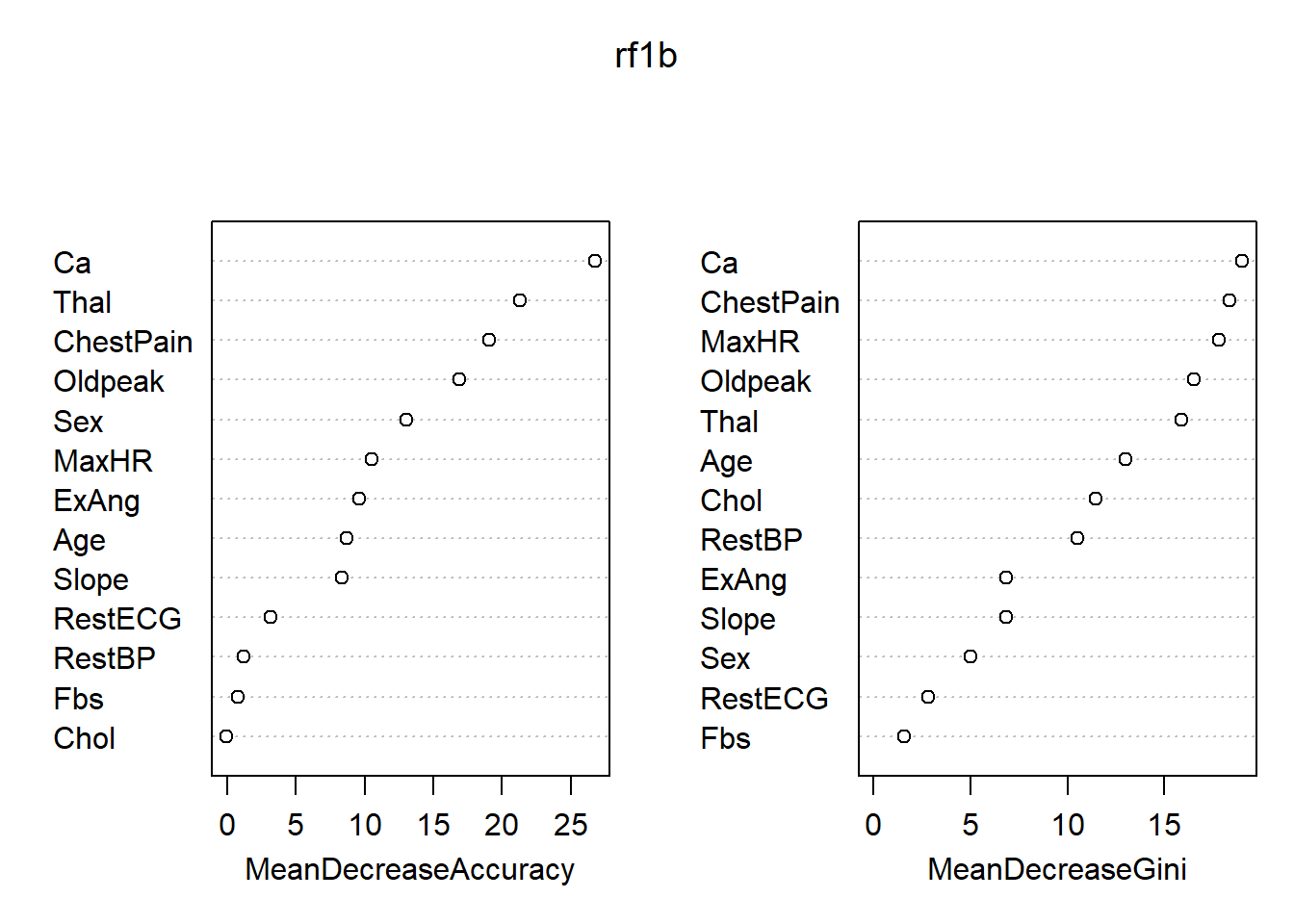
图43.4: Heart数据随机森林方法得到的变量重要度
最重要的变量是Ca, ChestPain。
43.7 附录
43.7.1 Heart数据
| Age | Sex | ChestPain | RestBP | Chol | Fbs | RestECG | MaxHR | ExAng | Oldpeak | Slope | Ca | Thal | AHD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 63 | 1 | typical | 145 | 233 | 1 | 2 | 150 | 0 | 2.3 | 3 | 0 | fixed | No |
| 67 | 1 | asymptomatic | 160 | 286 | 0 | 2 | 108 | 1 | 1.5 | 2 | 3 | normal | Yes |
| 67 | 1 | asymptomatic | 120 | 229 | 0 | 2 | 129 | 1 | 2.6 | 2 | 2 | reversable | Yes |
| 37 | 1 | nonanginal | 130 | 250 | 0 | 0 | 187 | 0 | 3.5 | 3 | 0 | normal | No |
| 41 | 0 | nontypical | 130 | 204 | 0 | 2 | 172 | 0 | 1.4 | 1 | 0 | normal | No |
| 56 | 1 | nontypical | 120 | 236 | 0 | 0 | 178 | 0 | 0.8 | 1 | 0 | normal | No |
| 62 | 0 | asymptomatic | 140 | 268 | 0 | 2 | 160 | 0 | 3.6 | 3 | 2 | normal | Yes |
| 57 | 0 | asymptomatic | 120 | 354 | 0 | 0 | 163 | 1 | 0.6 | 1 | 0 | normal | No |
| 63 | 1 | asymptomatic | 130 | 254 | 0 | 2 | 147 | 0 | 1.4 | 2 | 1 | reversable | Yes |
| 53 | 1 | asymptomatic | 140 | 203 | 1 | 2 | 155 | 1 | 3.1 | 3 | 0 | reversable | Yes |
| 57 | 1 | asymptomatic | 140 | 192 | 0 | 0 | 148 | 0 | 0.4 | 2 | 0 | fixed | No |
| 56 | 0 | nontypical | 140 | 294 | 0 | 2 | 153 | 0 | 1.3 | 2 | 0 | normal | No |
| 56 | 1 | nonanginal | 130 | 256 | 1 | 2 | 142 | 1 | 0.6 | 2 | 1 | fixed | Yes |
| 44 | 1 | nontypical | 120 | 263 | 0 | 0 | 173 | 0 | 0.0 | 1 | 0 | reversable | No |
| 52 | 1 | nonanginal | 172 | 199 | 1 | 0 | 162 | 0 | 0.5 | 1 | 0 | reversable | No |
| 57 | 1 | nonanginal | 150 | 168 | 0 | 0 | 174 | 0 | 1.6 | 1 | 0 | normal | No |
| 48 | 1 | nontypical | 110 | 229 | 0 | 0 | 168 | 0 | 1.0 | 3 | 0 | reversable | Yes |
| 54 | 1 | asymptomatic | 140 | 239 | 0 | 0 | 160 | 0 | 1.2 | 1 | 0 | normal | No |
| 48 | 0 | nonanginal | 130 | 275 | 0 | 0 | 139 | 0 | 0.2 | 1 | 0 | normal | No |
| 49 | 1 | nontypical | 130 | 266 | 0 | 0 | 171 | 0 | 0.6 | 1 | 0 | normal | No |
| 64 | 1 | typical | 110 | 211 | 0 | 2 | 144 | 1 | 1.8 | 2 | 0 | normal | No |
| 58 | 0 | typical | 150 | 283 | 1 | 2 | 162 | 0 | 1.0 | 1 | 0 | normal | No |
| 58 | 1 | nontypical | 120 | 284 | 0 | 2 | 160 | 0 | 1.8 | 2 | 0 | normal | Yes |
| 58 | 1 | nonanginal | 132 | 224 | 0 | 2 | 173 | 0 | 3.2 | 1 | 2 | reversable | Yes |
| 60 | 1 | asymptomatic | 130 | 206 | 0 | 2 | 132 | 1 | 2.4 | 2 | 2 | reversable | Yes |
| 50 | 0 | nonanginal | 120 | 219 | 0 | 0 | 158 | 0 | 1.6 | 2 | 0 | normal | No |
| 58 | 0 | nonanginal | 120 | 340 | 0 | 0 | 172 | 0 | 0.0 | 1 | 0 | normal | No |
| 66 | 0 | typical | 150 | 226 | 0 | 0 | 114 | 0 | 2.6 | 3 | 0 | normal | No |
| 43 | 1 | asymptomatic | 150 | 247 | 0 | 0 | 171 | 0 | 1.5 | 1 | 0 | normal | No |
| 40 | 1 | asymptomatic | 110 | 167 | 0 | 2 | 114 | 1 | 2.0 | 2 | 0 | reversable | Yes |
| 69 | 0 | typical | 140 | 239 | 0 | 0 | 151 | 0 | 1.8 | 1 | 2 | normal | No |
| 60 | 1 | asymptomatic | 117 | 230 | 1 | 0 | 160 | 1 | 1.4 | 1 | 2 | reversable | Yes |
| 64 | 1 | nonanginal | 140 | 335 | 0 | 0 | 158 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 59 | 1 | asymptomatic | 135 | 234 | 0 | 0 | 161 | 0 | 0.5 | 2 | 0 | reversable | No |
| 44 | 1 | nonanginal | 130 | 233 | 0 | 0 | 179 | 1 | 0.4 | 1 | 0 | normal | No |
| 42 | 1 | asymptomatic | 140 | 226 | 0 | 0 | 178 | 0 | 0.0 | 1 | 0 | normal | No |
| 43 | 1 | asymptomatic | 120 | 177 | 0 | 2 | 120 | 1 | 2.5 | 2 | 0 | reversable | Yes |
| 57 | 1 | asymptomatic | 150 | 276 | 0 | 2 | 112 | 1 | 0.6 | 2 | 1 | fixed | Yes |
| 55 | 1 | asymptomatic | 132 | 353 | 0 | 0 | 132 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 61 | 1 | nonanginal | 150 | 243 | 1 | 0 | 137 | 1 | 1.0 | 2 | 0 | normal | No |
| 65 | 0 | asymptomatic | 150 | 225 | 0 | 2 | 114 | 0 | 1.0 | 2 | 3 | reversable | Yes |
| 40 | 1 | typical | 140 | 199 | 0 | 0 | 178 | 1 | 1.4 | 1 | 0 | reversable | No |
| 71 | 0 | nontypical | 160 | 302 | 0 | 0 | 162 | 0 | 0.4 | 1 | 2 | normal | No |
| 59 | 1 | nonanginal | 150 | 212 | 1 | 0 | 157 | 0 | 1.6 | 1 | 0 | normal | No |
| 61 | 0 | asymptomatic | 130 | 330 | 0 | 2 | 169 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 58 | 1 | nonanginal | 112 | 230 | 0 | 2 | 165 | 0 | 2.5 | 2 | 1 | reversable | Yes |
| 51 | 1 | nonanginal | 110 | 175 | 0 | 0 | 123 | 0 | 0.6 | 1 | 0 | normal | No |
| 50 | 1 | asymptomatic | 150 | 243 | 0 | 2 | 128 | 0 | 2.6 | 2 | 0 | reversable | Yes |
| 65 | 0 | nonanginal | 140 | 417 | 1 | 2 | 157 | 0 | 0.8 | 1 | 1 | normal | No |
| 53 | 1 | nonanginal | 130 | 197 | 1 | 2 | 152 | 0 | 1.2 | 3 | 0 | normal | No |
| 41 | 0 | nontypical | 105 | 198 | 0 | 0 | 168 | 0 | 0.0 | 1 | 1 | normal | No |
| 65 | 1 | asymptomatic | 120 | 177 | 0 | 0 | 140 | 0 | 0.4 | 1 | 0 | reversable | No |
| 44 | 1 | asymptomatic | 112 | 290 | 0 | 2 | 153 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 44 | 1 | nontypical | 130 | 219 | 0 | 2 | 188 | 0 | 0.0 | 1 | 0 | normal | No |
| 60 | 1 | asymptomatic | 130 | 253 | 0 | 0 | 144 | 1 | 1.4 | 1 | 1 | reversable | Yes |
| 54 | 1 | asymptomatic | 124 | 266 | 0 | 2 | 109 | 1 | 2.2 | 2 | 1 | reversable | Yes |
| 50 | 1 | nonanginal | 140 | 233 | 0 | 0 | 163 | 0 | 0.6 | 2 | 1 | reversable | Yes |
| 41 | 1 | asymptomatic | 110 | 172 | 0 | 2 | 158 | 0 | 0.0 | 1 | 0 | reversable | Yes |
| 54 | 1 | nonanginal | 125 | 273 | 0 | 2 | 152 | 0 | 0.5 | 3 | 1 | normal | No |
| 51 | 1 | typical | 125 | 213 | 0 | 2 | 125 | 1 | 1.4 | 1 | 1 | normal | No |
| 51 | 0 | asymptomatic | 130 | 305 | 0 | 0 | 142 | 1 | 1.2 | 2 | 0 | reversable | Yes |
| 46 | 0 | nonanginal | 142 | 177 | 0 | 2 | 160 | 1 | 1.4 | 3 | 0 | normal | No |
| 58 | 1 | asymptomatic | 128 | 216 | 0 | 2 | 131 | 1 | 2.2 | 2 | 3 | reversable | Yes |
| 54 | 0 | nonanginal | 135 | 304 | 1 | 0 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 54 | 1 | asymptomatic | 120 | 188 | 0 | 0 | 113 | 0 | 1.4 | 2 | 1 | reversable | Yes |
| 60 | 1 | asymptomatic | 145 | 282 | 0 | 2 | 142 | 1 | 2.8 | 2 | 2 | reversable | Yes |
| 60 | 1 | nonanginal | 140 | 185 | 0 | 2 | 155 | 0 | 3.0 | 2 | 0 | normal | Yes |
| 54 | 1 | nonanginal | 150 | 232 | 0 | 2 | 165 | 0 | 1.6 | 1 | 0 | reversable | No |
| 59 | 1 | asymptomatic | 170 | 326 | 0 | 2 | 140 | 1 | 3.4 | 3 | 0 | reversable | Yes |
| 46 | 1 | nonanginal | 150 | 231 | 0 | 0 | 147 | 0 | 3.6 | 2 | 0 | normal | Yes |
| 65 | 0 | nonanginal | 155 | 269 | 0 | 0 | 148 | 0 | 0.8 | 1 | 0 | normal | No |
| 67 | 1 | asymptomatic | 125 | 254 | 1 | 0 | 163 | 0 | 0.2 | 2 | 2 | reversable | Yes |
| 62 | 1 | asymptomatic | 120 | 267 | 0 | 0 | 99 | 1 | 1.8 | 2 | 2 | reversable | Yes |
| 65 | 1 | asymptomatic | 110 | 248 | 0 | 2 | 158 | 0 | 0.6 | 1 | 2 | fixed | Yes |
| 44 | 1 | asymptomatic | 110 | 197 | 0 | 2 | 177 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 65 | 0 | nonanginal | 160 | 360 | 0 | 2 | 151 | 0 | 0.8 | 1 | 0 | normal | No |
| 60 | 1 | asymptomatic | 125 | 258 | 0 | 2 | 141 | 1 | 2.8 | 2 | 1 | reversable | Yes |
| 51 | 0 | nonanginal | 140 | 308 | 0 | 2 | 142 | 0 | 1.5 | 1 | 1 | normal | No |
| 48 | 1 | nontypical | 130 | 245 | 0 | 2 | 180 | 0 | 0.2 | 2 | 0 | normal | No |
| 58 | 1 | asymptomatic | 150 | 270 | 0 | 2 | 111 | 1 | 0.8 | 1 | 0 | reversable | Yes |
| 45 | 1 | asymptomatic | 104 | 208 | 0 | 2 | 148 | 1 | 3.0 | 2 | 0 | normal | No |
| 53 | 0 | asymptomatic | 130 | 264 | 0 | 2 | 143 | 0 | 0.4 | 2 | 0 | normal | No |
| 39 | 1 | nonanginal | 140 | 321 | 0 | 2 | 182 | 0 | 0.0 | 1 | 0 | normal | No |
| 68 | 1 | nonanginal | 180 | 274 | 1 | 2 | 150 | 1 | 1.6 | 2 | 0 | reversable | Yes |
| 52 | 1 | nontypical | 120 | 325 | 0 | 0 | 172 | 0 | 0.2 | 1 | 0 | normal | No |
| 44 | 1 | nonanginal | 140 | 235 | 0 | 2 | 180 | 0 | 0.0 | 1 | 0 | normal | No |
| 47 | 1 | nonanginal | 138 | 257 | 0 | 2 | 156 | 0 | 0.0 | 1 | 0 | normal | No |
| 53 | 0 | asymptomatic | 138 | 234 | 0 | 2 | 160 | 0 | 0.0 | 1 | 0 | normal | No |
| 51 | 0 | nonanginal | 130 | 256 | 0 | 2 | 149 | 0 | 0.5 | 1 | 0 | normal | No |
| 66 | 1 | asymptomatic | 120 | 302 | 0 | 2 | 151 | 0 | 0.4 | 2 | 0 | normal | No |
| 62 | 0 | asymptomatic | 160 | 164 | 0 | 2 | 145 | 0 | 6.2 | 3 | 3 | reversable | Yes |
| 62 | 1 | nonanginal | 130 | 231 | 0 | 0 | 146 | 0 | 1.8 | 2 | 3 | reversable | No |
| 44 | 0 | nonanginal | 108 | 141 | 0 | 0 | 175 | 0 | 0.6 | 2 | 0 | normal | No |
| 63 | 0 | nonanginal | 135 | 252 | 0 | 2 | 172 | 0 | 0.0 | 1 | 0 | normal | No |
| 52 | 1 | asymptomatic | 128 | 255 | 0 | 0 | 161 | 1 | 0.0 | 1 | 1 | reversable | Yes |
| 59 | 1 | asymptomatic | 110 | 239 | 0 | 2 | 142 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 60 | 0 | asymptomatic | 150 | 258 | 0 | 2 | 157 | 0 | 2.6 | 2 | 2 | reversable | Yes |
| 52 | 1 | nontypical | 134 | 201 | 0 | 0 | 158 | 0 | 0.8 | 1 | 1 | normal | No |
| 48 | 1 | asymptomatic | 122 | 222 | 0 | 2 | 186 | 0 | 0.0 | 1 | 0 | normal | No |
| 45 | 1 | asymptomatic | 115 | 260 | 0 | 2 | 185 | 0 | 0.0 | 1 | 0 | normal | No |
| 34 | 1 | typical | 118 | 182 | 0 | 2 | 174 | 0 | 0.0 | 1 | 0 | normal | No |
| 57 | 0 | asymptomatic | 128 | 303 | 0 | 2 | 159 | 0 | 0.0 | 1 | 1 | normal | No |
| 71 | 0 | nonanginal | 110 | 265 | 1 | 2 | 130 | 0 | 0.0 | 1 | 1 | normal | No |
| 49 | 1 | nonanginal | 120 | 188 | 0 | 0 | 139 | 0 | 2.0 | 2 | 3 | reversable | Yes |
| 54 | 1 | nontypical | 108 | 309 | 0 | 0 | 156 | 0 | 0.0 | 1 | 0 | reversable | No |
| 59 | 1 | asymptomatic | 140 | 177 | 0 | 0 | 162 | 1 | 0.0 | 1 | 1 | reversable | Yes |
| 57 | 1 | nonanginal | 128 | 229 | 0 | 2 | 150 | 0 | 0.4 | 2 | 1 | reversable | Yes |
| 61 | 1 | asymptomatic | 120 | 260 | 0 | 0 | 140 | 1 | 3.6 | 2 | 1 | reversable | Yes |
| 39 | 1 | asymptomatic | 118 | 219 | 0 | 0 | 140 | 0 | 1.2 | 2 | 0 | reversable | Yes |
| 61 | 0 | asymptomatic | 145 | 307 | 0 | 2 | 146 | 1 | 1.0 | 2 | 0 | reversable | Yes |
| 56 | 1 | asymptomatic | 125 | 249 | 1 | 2 | 144 | 1 | 1.2 | 2 | 1 | normal | Yes |
| 52 | 1 | typical | 118 | 186 | 0 | 2 | 190 | 0 | 0.0 | 2 | 0 | fixed | No |
| 43 | 0 | asymptomatic | 132 | 341 | 1 | 2 | 136 | 1 | 3.0 | 2 | 0 | reversable | Yes |
| 62 | 0 | nonanginal | 130 | 263 | 0 | 0 | 97 | 0 | 1.2 | 2 | 1 | reversable | Yes |
| 41 | 1 | nontypical | 135 | 203 | 0 | 0 | 132 | 0 | 0.0 | 2 | 0 | fixed | No |
| 58 | 1 | nonanginal | 140 | 211 | 1 | 2 | 165 | 0 | 0.0 | 1 | 0 | normal | No |
| 35 | 0 | asymptomatic | 138 | 183 | 0 | 0 | 182 | 0 | 1.4 | 1 | 0 | normal | No |
| 63 | 1 | asymptomatic | 130 | 330 | 1 | 2 | 132 | 1 | 1.8 | 1 | 3 | reversable | Yes |
| 65 | 1 | asymptomatic | 135 | 254 | 0 | 2 | 127 | 0 | 2.8 | 2 | 1 | reversable | Yes |
| 48 | 1 | asymptomatic | 130 | 256 | 1 | 2 | 150 | 1 | 0.0 | 1 | 2 | reversable | Yes |
| 63 | 0 | asymptomatic | 150 | 407 | 0 | 2 | 154 | 0 | 4.0 | 2 | 3 | reversable | Yes |
| 51 | 1 | nonanginal | 100 | 222 | 0 | 0 | 143 | 1 | 1.2 | 2 | 0 | normal | No |
| 55 | 1 | asymptomatic | 140 | 217 | 0 | 0 | 111 | 1 | 5.6 | 3 | 0 | reversable | Yes |
| 65 | 1 | typical | 138 | 282 | 1 | 2 | 174 | 0 | 1.4 | 2 | 1 | normal | Yes |
| 45 | 0 | nontypical | 130 | 234 | 0 | 2 | 175 | 0 | 0.6 | 2 | 0 | normal | No |
| 56 | 0 | asymptomatic | 200 | 288 | 1 | 2 | 133 | 1 | 4.0 | 3 | 2 | reversable | Yes |
| 54 | 1 | asymptomatic | 110 | 239 | 0 | 0 | 126 | 1 | 2.8 | 2 | 1 | reversable | Yes |
| 44 | 1 | nontypical | 120 | 220 | 0 | 0 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 62 | 0 | asymptomatic | 124 | 209 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 54 | 1 | nonanginal | 120 | 258 | 0 | 2 | 147 | 0 | 0.4 | 2 | 0 | reversable | No |
| 51 | 1 | nonanginal | 94 | 227 | 0 | 0 | 154 | 1 | 0.0 | 1 | 1 | reversable | No |
| 29 | 1 | nontypical | 130 | 204 | 0 | 2 | 202 | 0 | 0.0 | 1 | 0 | normal | No |
| 51 | 1 | asymptomatic | 140 | 261 | 0 | 2 | 186 | 1 | 0.0 | 1 | 0 | normal | No |
| 43 | 0 | nonanginal | 122 | 213 | 0 | 0 | 165 | 0 | 0.2 | 2 | 0 | normal | No |
| 55 | 0 | nontypical | 135 | 250 | 0 | 2 | 161 | 0 | 1.4 | 2 | 0 | normal | No |
| 70 | 1 | asymptomatic | 145 | 174 | 0 | 0 | 125 | 1 | 2.6 | 3 | 0 | reversable | Yes |
| 62 | 1 | nontypical | 120 | 281 | 0 | 2 | 103 | 0 | 1.4 | 2 | 1 | reversable | Yes |
| 35 | 1 | asymptomatic | 120 | 198 | 0 | 0 | 130 | 1 | 1.6 | 2 | 0 | reversable | Yes |
| 51 | 1 | nonanginal | 125 | 245 | 1 | 2 | 166 | 0 | 2.4 | 2 | 0 | normal | No |
| 59 | 1 | nontypical | 140 | 221 | 0 | 0 | 164 | 1 | 0.0 | 1 | 0 | normal | No |
| 59 | 1 | typical | 170 | 288 | 0 | 2 | 159 | 0 | 0.2 | 2 | 0 | reversable | Yes |
| 52 | 1 | nontypical | 128 | 205 | 1 | 0 | 184 | 0 | 0.0 | 1 | 0 | normal | No |
| 64 | 1 | nonanginal | 125 | 309 | 0 | 0 | 131 | 1 | 1.8 | 2 | 0 | reversable | Yes |
| 58 | 1 | nonanginal | 105 | 240 | 0 | 2 | 154 | 1 | 0.6 | 2 | 0 | reversable | No |
| 47 | 1 | nonanginal | 108 | 243 | 0 | 0 | 152 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 57 | 1 | asymptomatic | 165 | 289 | 1 | 2 | 124 | 0 | 1.0 | 2 | 3 | reversable | Yes |
| 41 | 1 | nonanginal | 112 | 250 | 0 | 0 | 179 | 0 | 0.0 | 1 | 0 | normal | No |
| 45 | 1 | nontypical | 128 | 308 | 0 | 2 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 60 | 0 | nonanginal | 102 | 318 | 0 | 0 | 160 | 0 | 0.0 | 1 | 1 | normal | No |
| 52 | 1 | typical | 152 | 298 | 1 | 0 | 178 | 0 | 1.2 | 2 | 0 | reversable | No |
| 42 | 0 | asymptomatic | 102 | 265 | 0 | 2 | 122 | 0 | 0.6 | 2 | 0 | normal | No |
| 67 | 0 | nonanginal | 115 | 564 | 0 | 2 | 160 | 0 | 1.6 | 2 | 0 | reversable | No |
| 55 | 1 | asymptomatic | 160 | 289 | 0 | 2 | 145 | 1 | 0.8 | 2 | 1 | reversable | Yes |
| 64 | 1 | asymptomatic | 120 | 246 | 0 | 2 | 96 | 1 | 2.2 | 3 | 1 | normal | Yes |
| 70 | 1 | asymptomatic | 130 | 322 | 0 | 2 | 109 | 0 | 2.4 | 2 | 3 | normal | Yes |
| 51 | 1 | asymptomatic | 140 | 299 | 0 | 0 | 173 | 1 | 1.6 | 1 | 0 | reversable | Yes |
| 58 | 1 | asymptomatic | 125 | 300 | 0 | 2 | 171 | 0 | 0.0 | 1 | 2 | reversable | Yes |
| 60 | 1 | asymptomatic | 140 | 293 | 0 | 2 | 170 | 0 | 1.2 | 2 | 2 | reversable | Yes |
| 68 | 1 | nonanginal | 118 | 277 | 0 | 0 | 151 | 0 | 1.0 | 1 | 1 | reversable | No |
| 46 | 1 | nontypical | 101 | 197 | 1 | 0 | 156 | 0 | 0.0 | 1 | 0 | reversable | No |
| 77 | 1 | asymptomatic | 125 | 304 | 0 | 2 | 162 | 1 | 0.0 | 1 | 3 | normal | Yes |
| 54 | 0 | nonanginal | 110 | 214 | 0 | 0 | 158 | 0 | 1.6 | 2 | 0 | normal | No |
| 58 | 0 | asymptomatic | 100 | 248 | 0 | 2 | 122 | 0 | 1.0 | 2 | 0 | normal | No |
| 48 | 1 | nonanginal | 124 | 255 | 1 | 0 | 175 | 0 | 0.0 | 1 | 2 | normal | No |
| 57 | 1 | asymptomatic | 132 | 207 | 0 | 0 | 168 | 1 | 0.0 | 1 | 0 | reversable | No |
| 54 | 0 | nontypical | 132 | 288 | 1 | 2 | 159 | 1 | 0.0 | 1 | 1 | normal | No |
| 35 | 1 | asymptomatic | 126 | 282 | 0 | 2 | 156 | 1 | 0.0 | 1 | 0 | reversable | Yes |
| 45 | 0 | nontypical | 112 | 160 | 0 | 0 | 138 | 0 | 0.0 | 2 | 0 | normal | No |
| 70 | 1 | nonanginal | 160 | 269 | 0 | 0 | 112 | 1 | 2.9 | 2 | 1 | reversable | Yes |
| 53 | 1 | asymptomatic | 142 | 226 | 0 | 2 | 111 | 1 | 0.0 | 1 | 0 | reversable | No |
| 59 | 0 | asymptomatic | 174 | 249 | 0 | 0 | 143 | 1 | 0.0 | 2 | 0 | normal | Yes |
| 62 | 0 | asymptomatic | 140 | 394 | 0 | 2 | 157 | 0 | 1.2 | 2 | 0 | normal | No |
| 64 | 1 | asymptomatic | 145 | 212 | 0 | 2 | 132 | 0 | 2.0 | 2 | 2 | fixed | Yes |
| 57 | 1 | asymptomatic | 152 | 274 | 0 | 0 | 88 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 52 | 1 | asymptomatic | 108 | 233 | 1 | 0 | 147 | 0 | 0.1 | 1 | 3 | reversable | No |
| 56 | 1 | asymptomatic | 132 | 184 | 0 | 2 | 105 | 1 | 2.1 | 2 | 1 | fixed | Yes |
| 43 | 1 | nonanginal | 130 | 315 | 0 | 0 | 162 | 0 | 1.9 | 1 | 1 | normal | No |
| 53 | 1 | nonanginal | 130 | 246 | 1 | 2 | 173 | 0 | 0.0 | 1 | 3 | normal | No |
| 48 | 1 | asymptomatic | 124 | 274 | 0 | 2 | 166 | 0 | 0.5 | 2 | 0 | reversable | Yes |
| 56 | 0 | asymptomatic | 134 | 409 | 0 | 2 | 150 | 1 | 1.9 | 2 | 2 | reversable | Yes |
| 42 | 1 | typical | 148 | 244 | 0 | 2 | 178 | 0 | 0.8 | 1 | 2 | normal | No |
| 59 | 1 | typical | 178 | 270 | 0 | 2 | 145 | 0 | 4.2 | 3 | 0 | reversable | No |
| 60 | 0 | asymptomatic | 158 | 305 | 0 | 2 | 161 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 63 | 0 | nontypical | 140 | 195 | 0 | 0 | 179 | 0 | 0.0 | 1 | 2 | normal | No |
| 42 | 1 | nonanginal | 120 | 240 | 1 | 0 | 194 | 0 | 0.8 | 3 | 0 | reversable | No |
| 66 | 1 | nontypical | 160 | 246 | 0 | 0 | 120 | 1 | 0.0 | 2 | 3 | fixed | Yes |
| 54 | 1 | nontypical | 192 | 283 | 0 | 2 | 195 | 0 | 0.0 | 1 | 1 | reversable | Yes |
| 69 | 1 | nonanginal | 140 | 254 | 0 | 2 | 146 | 0 | 2.0 | 2 | 3 | reversable | Yes |
| 50 | 1 | nonanginal | 129 | 196 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 51 | 1 | asymptomatic | 140 | 298 | 0 | 0 | 122 | 1 | 4.2 | 2 | 3 | reversable | Yes |
| 62 | 0 | asymptomatic | 138 | 294 | 1 | 0 | 106 | 0 | 1.9 | 2 | 3 | normal | Yes |
| 68 | 0 | nonanginal | 120 | 211 | 0 | 2 | 115 | 0 | 1.5 | 2 | 0 | normal | No |
| 67 | 1 | asymptomatic | 100 | 299 | 0 | 2 | 125 | 1 | 0.9 | 2 | 2 | normal | Yes |
| 69 | 1 | typical | 160 | 234 | 1 | 2 | 131 | 0 | 0.1 | 2 | 1 | normal | No |
| 45 | 0 | asymptomatic | 138 | 236 | 0 | 2 | 152 | 1 | 0.2 | 2 | 0 | normal | No |
| 50 | 0 | nontypical | 120 | 244 | 0 | 0 | 162 | 0 | 1.1 | 1 | 0 | normal | No |
| 59 | 1 | typical | 160 | 273 | 0 | 2 | 125 | 0 | 0.0 | 1 | 0 | normal | Yes |
| 50 | 0 | asymptomatic | 110 | 254 | 0 | 2 | 159 | 0 | 0.0 | 1 | 0 | normal | No |
| 64 | 0 | asymptomatic | 180 | 325 | 0 | 0 | 154 | 1 | 0.0 | 1 | 0 | normal | No |
| 57 | 1 | nonanginal | 150 | 126 | 1 | 0 | 173 | 0 | 0.2 | 1 | 1 | reversable | No |
| 64 | 0 | nonanginal | 140 | 313 | 0 | 0 | 133 | 0 | 0.2 | 1 | 0 | reversable | No |
| 43 | 1 | asymptomatic | 110 | 211 | 0 | 0 | 161 | 0 | 0.0 | 1 | 0 | reversable | No |
| 45 | 1 | asymptomatic | 142 | 309 | 0 | 2 | 147 | 1 | 0.0 | 2 | 3 | reversable | Yes |
| 58 | 1 | asymptomatic | 128 | 259 | 0 | 2 | 130 | 1 | 3.0 | 2 | 2 | reversable | Yes |
| 50 | 1 | asymptomatic | 144 | 200 | 0 | 2 | 126 | 1 | 0.9 | 2 | 0 | reversable | Yes |
| 55 | 1 | nontypical | 130 | 262 | 0 | 0 | 155 | 0 | 0.0 | 1 | 0 | normal | No |
| 62 | 0 | asymptomatic | 150 | 244 | 0 | 0 | 154 | 1 | 1.4 | 2 | 0 | normal | Yes |
| 37 | 0 | nonanginal | 120 | 215 | 0 | 0 | 170 | 0 | 0.0 | 1 | 0 | normal | No |
| 38 | 1 | typical | 120 | 231 | 0 | 0 | 182 | 1 | 3.8 | 2 | 0 | reversable | Yes |
| 41 | 1 | nonanginal | 130 | 214 | 0 | 2 | 168 | 0 | 2.0 | 2 | 0 | normal | No |
| 66 | 0 | asymptomatic | 178 | 228 | 1 | 0 | 165 | 1 | 1.0 | 2 | 2 | reversable | Yes |
| 52 | 1 | asymptomatic | 112 | 230 | 0 | 0 | 160 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 56 | 1 | typical | 120 | 193 | 0 | 2 | 162 | 0 | 1.9 | 2 | 0 | reversable | No |
| 46 | 0 | nontypical | 105 | 204 | 0 | 0 | 172 | 0 | 0.0 | 1 | 0 | normal | No |
| 46 | 0 | asymptomatic | 138 | 243 | 0 | 2 | 152 | 1 | 0.0 | 2 | 0 | normal | No |
| 64 | 0 | asymptomatic | 130 | 303 | 0 | 0 | 122 | 0 | 2.0 | 2 | 2 | normal | No |
| 59 | 1 | asymptomatic | 138 | 271 | 0 | 2 | 182 | 0 | 0.0 | 1 | 0 | normal | No |
| 41 | 0 | nonanginal | 112 | 268 | 0 | 2 | 172 | 1 | 0.0 | 1 | 0 | normal | No |
| 54 | 0 | nonanginal | 108 | 267 | 0 | 2 | 167 | 0 | 0.0 | 1 | 0 | normal | No |
| 39 | 0 | nonanginal | 94 | 199 | 0 | 0 | 179 | 0 | 0.0 | 1 | 0 | normal | No |
| 53 | 1 | asymptomatic | 123 | 282 | 0 | 0 | 95 | 1 | 2.0 | 2 | 2 | reversable | Yes |
| 63 | 0 | asymptomatic | 108 | 269 | 0 | 0 | 169 | 1 | 1.8 | 2 | 2 | normal | Yes |
| 34 | 0 | nontypical | 118 | 210 | 0 | 0 | 192 | 0 | 0.7 | 1 | 0 | normal | No |
| 47 | 1 | asymptomatic | 112 | 204 | 0 | 0 | 143 | 0 | 0.1 | 1 | 0 | normal | No |
| 67 | 0 | nonanginal | 152 | 277 | 0 | 0 | 172 | 0 | 0.0 | 1 | 1 | normal | No |
| 54 | 1 | asymptomatic | 110 | 206 | 0 | 2 | 108 | 1 | 0.0 | 2 | 1 | normal | Yes |
| 66 | 1 | asymptomatic | 112 | 212 | 0 | 2 | 132 | 1 | 0.1 | 1 | 1 | normal | Yes |
| 52 | 0 | nonanginal | 136 | 196 | 0 | 2 | 169 | 0 | 0.1 | 2 | 0 | normal | No |
| 55 | 0 | asymptomatic | 180 | 327 | 0 | 1 | 117 | 1 | 3.4 | 2 | 0 | normal | Yes |
| 49 | 1 | nonanginal | 118 | 149 | 0 | 2 | 126 | 0 | 0.8 | 1 | 3 | normal | Yes |
| 74 | 0 | nontypical | 120 | 269 | 0 | 2 | 121 | 1 | 0.2 | 1 | 1 | normal | No |
| 54 | 0 | nonanginal | 160 | 201 | 0 | 0 | 163 | 0 | 0.0 | 1 | 1 | normal | No |
| 54 | 1 | asymptomatic | 122 | 286 | 0 | 2 | 116 | 1 | 3.2 | 2 | 2 | normal | Yes |
| 56 | 1 | asymptomatic | 130 | 283 | 1 | 2 | 103 | 1 | 1.6 | 3 | 0 | reversable | Yes |
| 46 | 1 | asymptomatic | 120 | 249 | 0 | 2 | 144 | 0 | 0.8 | 1 | 0 | reversable | Yes |
| 49 | 0 | nontypical | 134 | 271 | 0 | 0 | 162 | 0 | 0.0 | 2 | 0 | normal | No |
| 42 | 1 | nontypical | 120 | 295 | 0 | 0 | 162 | 0 | 0.0 | 1 | 0 | normal | No |
| 41 | 1 | nontypical | 110 | 235 | 0 | 0 | 153 | 0 | 0.0 | 1 | 0 | normal | No |
| 41 | 0 | nontypical | 126 | 306 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 49 | 0 | asymptomatic | 130 | 269 | 0 | 0 | 163 | 0 | 0.0 | 1 | 0 | normal | No |
| 61 | 1 | typical | 134 | 234 | 0 | 0 | 145 | 0 | 2.6 | 2 | 2 | normal | Yes |
| 60 | 0 | nonanginal | 120 | 178 | 1 | 0 | 96 | 0 | 0.0 | 1 | 0 | normal | No |
| 67 | 1 | asymptomatic | 120 | 237 | 0 | 0 | 71 | 0 | 1.0 | 2 | 0 | normal | Yes |
| 58 | 1 | asymptomatic | 100 | 234 | 0 | 0 | 156 | 0 | 0.1 | 1 | 1 | reversable | Yes |
| 47 | 1 | asymptomatic | 110 | 275 | 0 | 2 | 118 | 1 | 1.0 | 2 | 1 | normal | Yes |
| 52 | 1 | asymptomatic | 125 | 212 | 0 | 0 | 168 | 0 | 1.0 | 1 | 2 | reversable | Yes |
| 62 | 1 | nontypical | 128 | 208 | 1 | 2 | 140 | 0 | 0.0 | 1 | 0 | normal | No |
| 57 | 1 | asymptomatic | 110 | 201 | 0 | 0 | 126 | 1 | 1.5 | 2 | 0 | fixed | No |
| 58 | 1 | asymptomatic | 146 | 218 | 0 | 0 | 105 | 0 | 2.0 | 2 | 1 | reversable | Yes |
| 64 | 1 | asymptomatic | 128 | 263 | 0 | 0 | 105 | 1 | 0.2 | 2 | 1 | reversable | No |
| 51 | 0 | nonanginal | 120 | 295 | 0 | 2 | 157 | 0 | 0.6 | 1 | 0 | normal | No |
| 43 | 1 | asymptomatic | 115 | 303 | 0 | 0 | 181 | 0 | 1.2 | 2 | 0 | normal | No |
| 42 | 0 | nonanginal | 120 | 209 | 0 | 0 | 173 | 0 | 0.0 | 2 | 0 | normal | No |
| 67 | 0 | asymptomatic | 106 | 223 | 0 | 0 | 142 | 0 | 0.3 | 1 | 2 | normal | No |
| 76 | 0 | nonanginal | 140 | 197 | 0 | 1 | 116 | 0 | 1.1 | 2 | 0 | normal | No |
| 70 | 1 | nontypical | 156 | 245 | 0 | 2 | 143 | 0 | 0.0 | 1 | 0 | normal | No |
| 57 | 1 | nontypical | 124 | 261 | 0 | 0 | 141 | 0 | 0.3 | 1 | 0 | reversable | Yes |
| 44 | 0 | nonanginal | 118 | 242 | 0 | 0 | 149 | 0 | 0.3 | 2 | 1 | normal | No |
| 58 | 0 | nontypical | 136 | 319 | 1 | 2 | 152 | 0 | 0.0 | 1 | 2 | normal | Yes |
| 60 | 0 | typical | 150 | 240 | 0 | 0 | 171 | 0 | 0.9 | 1 | 0 | normal | No |
| 44 | 1 | nonanginal | 120 | 226 | 0 | 0 | 169 | 0 | 0.0 | 1 | 0 | normal | No |
| 61 | 1 | asymptomatic | 138 | 166 | 0 | 2 | 125 | 1 | 3.6 | 2 | 1 | normal | Yes |
| 42 | 1 | asymptomatic | 136 | 315 | 0 | 0 | 125 | 1 | 1.8 | 2 | 0 | fixed | Yes |
| 59 | 1 | nonanginal | 126 | 218 | 1 | 0 | 134 | 0 | 2.2 | 2 | 1 | fixed | Yes |
| 40 | 1 | asymptomatic | 152 | 223 | 0 | 0 | 181 | 0 | 0.0 | 1 | 0 | reversable | Yes |
| 42 | 1 | nonanginal | 130 | 180 | 0 | 0 | 150 | 0 | 0.0 | 1 | 0 | normal | No |
| 61 | 1 | asymptomatic | 140 | 207 | 0 | 2 | 138 | 1 | 1.9 | 1 | 1 | reversable | Yes |
| 66 | 1 | asymptomatic | 160 | 228 | 0 | 2 | 138 | 0 | 2.3 | 1 | 0 | fixed | No |
| 46 | 1 | asymptomatic | 140 | 311 | 0 | 0 | 120 | 1 | 1.8 | 2 | 2 | reversable | Yes |
| 71 | 0 | asymptomatic | 112 | 149 | 0 | 0 | 125 | 0 | 1.6 | 2 | 0 | normal | No |
| 59 | 1 | typical | 134 | 204 | 0 | 0 | 162 | 0 | 0.8 | 1 | 2 | normal | Yes |
| 64 | 1 | typical | 170 | 227 | 0 | 2 | 155 | 0 | 0.6 | 2 | 0 | reversable | No |
| 66 | 0 | nonanginal | 146 | 278 | 0 | 2 | 152 | 0 | 0.0 | 2 | 1 | normal | No |
| 39 | 0 | nonanginal | 138 | 220 | 0 | 0 | 152 | 0 | 0.0 | 2 | 0 | normal | No |
| 57 | 1 | nontypical | 154 | 232 | 0 | 2 | 164 | 0 | 0.0 | 1 | 1 | normal | Yes |
| 58 | 0 | asymptomatic | 130 | 197 | 0 | 0 | 131 | 0 | 0.6 | 2 | 0 | normal | No |
| 57 | 1 | asymptomatic | 110 | 335 | 0 | 0 | 143 | 1 | 3.0 | 2 | 1 | reversable | Yes |
| 47 | 1 | nonanginal | 130 | 253 | 0 | 0 | 179 | 0 | 0.0 | 1 | 0 | normal | No |
| 55 | 0 | asymptomatic | 128 | 205 | 0 | 1 | 130 | 1 | 2.0 | 2 | 1 | reversable | Yes |
| 35 | 1 | nontypical | 122 | 192 | 0 | 0 | 174 | 0 | 0.0 | 1 | 0 | normal | No |
| 61 | 1 | asymptomatic | 148 | 203 | 0 | 0 | 161 | 0 | 0.0 | 1 | 1 | reversable | Yes |
| 58 | 1 | asymptomatic | 114 | 318 | 0 | 1 | 140 | 0 | 4.4 | 3 | 3 | fixed | Yes |
| 58 | 0 | asymptomatic | 170 | 225 | 1 | 2 | 146 | 1 | 2.8 | 2 | 2 | fixed | Yes |
| 56 | 1 | nontypical | 130 | 221 | 0 | 2 | 163 | 0 | 0.0 | 1 | 0 | reversable | No |
| 56 | 1 | nontypical | 120 | 240 | 0 | 0 | 169 | 0 | 0.0 | 3 | 0 | normal | No |
| 67 | 1 | nonanginal | 152 | 212 | 0 | 2 | 150 | 0 | 0.8 | 2 | 0 | reversable | Yes |
| 55 | 0 | nontypical | 132 | 342 | 0 | 0 | 166 | 0 | 1.2 | 1 | 0 | normal | No |
| 44 | 1 | asymptomatic | 120 | 169 | 0 | 0 | 144 | 1 | 2.8 | 3 | 0 | fixed | Yes |
| 63 | 1 | asymptomatic | 140 | 187 | 0 | 2 | 144 | 1 | 4.0 | 1 | 2 | reversable | Yes |
| 63 | 0 | asymptomatic | 124 | 197 | 0 | 0 | 136 | 1 | 0.0 | 2 | 0 | normal | Yes |
| 41 | 1 | nontypical | 120 | 157 | 0 | 0 | 182 | 0 | 0.0 | 1 | 0 | normal | No |
| 59 | 1 | asymptomatic | 164 | 176 | 1 | 2 | 90 | 0 | 1.0 | 2 | 2 | fixed | Yes |
| 57 | 0 | asymptomatic | 140 | 241 | 0 | 0 | 123 | 1 | 0.2 | 2 | 0 | reversable | Yes |
| 45 | 1 | typical | 110 | 264 | 0 | 0 | 132 | 0 | 1.2 | 2 | 0 | reversable | Yes |
| 68 | 1 | asymptomatic | 144 | 193 | 1 | 0 | 141 | 0 | 3.4 | 2 | 2 | reversable | Yes |
| 57 | 1 | asymptomatic | 130 | 131 | 0 | 0 | 115 | 1 | 1.2 | 2 | 1 | reversable | Yes |
| 57 | 0 | nontypical | 130 | 236 | 0 | 2 | 174 | 0 | 0.0 | 2 | 1 | normal | Yes |